14 - Aventuras na Covariâncias¶
Imports, loadings and functions¶
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
from matplotlib.patches import Ellipse
import matplotlib.transforms as transforms
import pandas as pd
import networkx as nx
# from causalgraphicalmodels import CausalGraphicalModel
import arviz as az
# ArviZ ships with style sheets!
# https://python.arviz.org/en/stable/examples/styles.html#example-styles
az.style.use("arviz-darkgrid")
import xarray as xr
import stan
import nest_asyncio
plt.style.use('default')
plt.rcParams['axes.facecolor'] = 'lightgray'
# To DAG's
import daft
from causalgraphicalmodels import CausalGraphicalModel # Just work in < python3.9
# Add fonts to matplotlib to run xkcd
from matplotlib import font_manager
font_dirs = ["fonts/"] # The path to the custom font file.
font_files = font_manager.findSystemFonts(fontpaths=font_dirs)
for font_file in font_files:
font_manager.fontManager.addfont(font_file)
# To make plots like drawing
# plt.xkcd()
# To running the stan in jupyter notebook
nest_asyncio.apply()
# https://matplotlib.org/stable/gallery/statistics/confidence_ellipse.html
def get_correlated_dataset(n, dependency, mu, scale):
latent = np.random.randn(n, 2)
dependent = latent.dot(dependency)
scaled = dependent * scale
scaled_with_offset = scaled + mu
# return x and y of the new, correlated dataset
return scaled_with_offset[:, 0], scaled_with_offset[:, 1]
# https://matplotlib.org/stable/gallery/statistics/confidence_ellipse.html
def confidence_ellipse(x, y, ax, n_std=3.0, facecolor='none', **kwargs):
"""
Create a plot of the covariance confidence ellipse of *x* and *y*.
Parameters
----------
x, y : array-like, shape (n, )
Input data.
ax : matplotlib.axes.Axes
The axes object to draw the ellipse into.
n_std : float
The number of standard deviations to determine the ellipse's radiuses.
**kwargs
Forwarded to `~matplotlib.patches.Ellipse`
Returns
-------
matplotlib.patches.Ellipse
"""
if x.size != y.size:
raise ValueError("x and y must be the same size")
cov = np.cov(x, y)
pearson = cov[0, 1]/np.sqrt(cov[0, 0] * cov[1, 1])
# Using a special case to obtain the eigenvalues of this
# two-dimensional dataset.
ell_radius_x = np.sqrt(1 + pearson)
ell_radius_y = np.sqrt(1 - pearson)
ellipse = Ellipse((0, 0), width=ell_radius_x * 2, height=ell_radius_y * 2,
facecolor=facecolor, **kwargs)
# Calculating the standard deviation of x from
# the squareroot of the variance and multiplying
# with the given number of standard deviations.
scale_x = np.sqrt(cov[0, 0]) * n_std
#scale_x = n_std
mean_x = np.mean(x)
# calculating the standard deviation of y ...
scale_y = np.sqrt(cov[1, 1]) * n_std
#scale_y = n_std
mean_y = np.mean(y)
transf = transforms.Affine2D() \
.rotate_deg(45) \
.scale(scale_x, scale_y) \
.translate(mean_x, mean_y)
ellipse.set_transform(transf + ax.transData)
return ax.add_patch(ellipse)
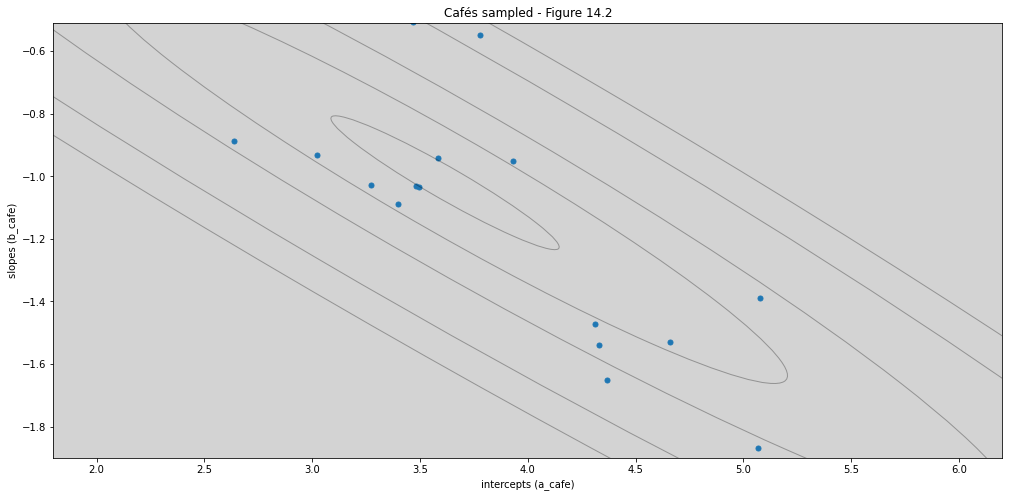
def draw_ellipse(mu, sigma, level, plot_points=None):
fig, ax_nstd = plt.subplots(figsize=(17, 8))
dependency_nstd = sigma.tolist()
scale = 5, 5
x, y = get_correlated_dataset(500, dependency_nstd, mu, scale)
if plot_points is not None:
ax_nstd.scatter(plot_points[:, 0], plot_points[:, 1], s=25)
for l in level:
confidence_ellipse(x, y, ax_nstd, n_std=l, edgecolor='black', alpha=0.3)
# ax_nstd.scatter(mu[0], mu[1], c='red', s=3)
ax_nstd.set_title('Cafés sampled - Figure 14.2')
ax_nstd.set_xlabel('intercepts (a_cafe)')
ax_nstd.set_ylabel('slopes (b_cafe)')
ax_nstd.set_xlim(1.8, 6.2)
ax_nstd.set_ylim(-1.9, -0.51)
plt.show()
14.1 Varing Slopes by Contructions¶
R Code 14.1¶
a = 3.5 # Average morning wait time
b = (-1) # Average difference afternoon wait time
sigma_a = 1 # std dev in intercepts
sigma_b = 0.5 # std in slopes
rho = (-0.7) # correlation between intercepts and slopes
R Code 14.2¶
Mu = np.array([a, b]) # Vector
Mu
array([ 3.5, -1. ])
R Code 14.3¶
The matrix variance and covariance:
And in math form:
R Code 14.3¶
cov_ab = sigma_a * sigma_b * rho
Sigma = np.matrix([
[sigma_a**2, cov_ab],
[cov_ab, sigma_b**2]
])
Sigma
matrix([[ 1. , -0.35],
[-0.35, 0.25]])
R Code 14.4¶
np.matrix([[1,2], [3, 4]])
matrix([[1, 2],
[3, 4]])
R Code 14.5¶
sigmas = np.array([sigma_a, sigma_b]) # Standart Deviations
sigmas
array([1. , 0.5])
Rho = np.matrix([
[1, rho],
[rho, 1]
]) # Correlation Matrix
Rho
matrix([[ 1. , -0.7],
[-0.7, 1. ]])
# Multiply to get covariance matrix
Sigma = np.diag(sigmas) * Rho * np.diag(sigmas)
Sigma
matrix([[ 1. , -0.35],
[-0.35, 0.25]])
R Code 14.6¶
N_cafe = 20
R Code 14.7¶
np.random.seed(5) # used to replicate example
vary_effects = stats.multivariate_normal(mean=Mu, cov=Sigma).rvs(N_cafe)
vary_effects
array([[ 3.02149593, -0.9304358 ],
[ 1.05666673, -0.12762876],
[ 3.58546284, -0.46432203],
[ 4.3297356 , -1.53991745],
[ 3.27332508, -1.02933771],
[ 4.65859716, -1.5303893 ],
[ 3.93018946, -0.9524405 ],
[ 5.06625508, -1.869279 ],
[ 2.58529637, 0.0287956 ],
[ 5.07890482, -1.39033508],
[ 4.36801698, -1.65042937],
[ 4.31342478, -1.47263713],
[ 2.59854231, -0.38835499],
[ 3.39670791, -1.09000246],
[ 3.48373345, -1.03170127],
[ 2.63542057, -0.88644913],
[ 3.49374238, -1.03389681],
[ 3.58250316, -0.94285315],
[ 3.46768242, -0.50699357],
[ 3.77789108, -0.54770738]])
R Code 14.8¶
a_cafe = vary_effects[:, 0] # Intercepts samples
b_cafe = vary_effects[:, 1] # Slopes samples
R Code 14.9¶
levels = [0.1, 0.3, 0.5, 0.8, 0.99]
draw_ellipse(mu=Mu, sigma=Sigma, level=levels, plot_points=vary_effects)
Overthinking: We define the covariance with 3 parameters: (pag 440)
Firt variable standart deviations \(\sigma_\alpha\)
Second variable standart deviations \( \sigma_\beta \)
Correlation between two variables \(\rho_{\alpha\beta}\)
Whu the covariance is equal to \(\sigma_\alpha \sigma_\beta \rho_{\alpha\beta}\)?
The usual definition of the covariance between two variables \(x\) and \(y\) is:
To simple variance this concept is the same:
If the variable with only mean equal zero (\(0\)), then the variance to be:
The correlation live in \([-1, 1]\), this is only the covariance rescaled. To be rescaled covariance just:
or better:
R Code 14.10¶
N_visit = 10
afternoon = np.tile([0, 1], reps=int((N_visit*N_cafe)/2))
cafe_id = np.repeat(np.arange(0, N_cafe), N_visit)
mu = a_cafe[cafe_id] + b_cafe[cafe_id]*afternoon
sigma = 0.5 # std dev within cafes
wait = np.random.normal(mu, sigma, size=N_visit*N_cafe)
cafe_id += 1 # index from 1 to 20 cafe_id, need to Stan index
d = pd.DataFrame({'cafe': cafe_id, 'afternoon':afternoon, 'wait': wait})
d
| cafe | afternoon | wait | |
|---|---|---|---|
| 0 | 1 | 0 | 2.713830 |
| 1 | 1 | 1 | 1.965617 |
| 2 | 1 | 0 | 3.261427 |
| 3 | 1 | 1 | 2.836598 |
| 4 | 1 | 0 | 3.105162 |
| ... | ... | ... | ... |
| 195 | 20 | 1 | 4.001264 |
| 196 | 20 | 0 | 3.991292 |
| 197 | 20 | 1 | 2.764103 |
| 198 | 20 | 0 | 3.507337 |
| 199 | 20 | 1 | 2.439118 |
200 rows × 3 columns
14.1.3 The varing slope model¶
The likelihood:
and the linear model:
Then comes the matrix of varing intercepts and slopes, with it’s covariance matrix:
Population of varying effects: $$
\sim Normal
The \(R\) is the correlation matrix. In this simple case \(R\) is defined like:
And the priors and hyper-priors:
prior for intercept:
prior for average slope:
prior for stddev within cafés
prior for stddev among cafés
prior for stddev among slopes
prior for correlation matrix
R Code 14.11¶
# Dont have the function LKJcorr implemented in numpy or scipy
# Stan implementation - https://mc-stan.org/docs/functions-reference/lkj-correlation.html
R Code 14.12¶
# https://mc-stan.org/docs/stan-users-guide/multivariate-hierarchical-priors.html
model = """
data {
int N;
int N_cafe;
int N_periods;
array[N] real wait;
array[N] int cafe;
array[N] int afternoon;
}
parameters {
real<lower=0> sigma;
vector<lower=0>[2] sigma_cafe;
vector[N_cafe] alpha_cafe;
vector[N_cafe] beta_cafe;
real alpha; // Like bar_alpha (hyper-prior)
real beta; // Like bar_beta (hyper-prior)
corr_matrix[N_periods] Rho;
}
model {
vector[N] mu;
vector[2] Y_cafe[N_cafe];
vector[2] MU_cafe;
//(sigma) - Prior to likelihood
sigma ~ exponential(1);
// (sigma_cafe) - Hyper Prior
sigma_cafe ~ exponential(1);
// (Rho) - Hyper Prior
Rho ~ lkj_corr(2);
// (MU_cafe) - Hyper Prior
alpha ~ normal(5, 2);
beta ~ normal(-1, 0.5);
MU_cafe = [alpha, beta]';
// (alpha_cafe, beta_cafe) - Prior
for (j in 1:N_cafe){
Y_cafe[j] = [alpha_cafe[j], beta_cafe[j]]' ;
}
Y_cafe ~ multi_normal(MU_cafe, quad_form_diag(Rho, sigma_cafe)); // Prior to alpha_cafe and beta_cafe
// (mu) - Linear model
for (i in 1:N){
mu[i] = alpha_cafe[ cafe[i] ] + beta_cafe[ cafe[i] ] * afternoon[i];
}
// Likelihood
wait ~ normal(mu, sigma);
}
"""
dat_list = {
'N': len(d),
'N_cafe': N_cafe,
'N_periods': len(d.afternoon.unique()),
'wait': d.wait.values,
'afternoon': d.afternoon.values,
'cafe': d.cafe.values,
}
posteriori = stan.build(model, data=dat_list)
samples = posteriori.sample(num_chains=4, num_samples=1000)
model_another_way = """
data {
int N;
int N_cafe;
int N_periods;
array[N] real wait;
array[N] int cafe;
array[N] int afternoon;
}
parameters {
real<lower=0> sigma;
vector<lower=0>[2] sigma_cafe;
vector[N_cafe] alpha_cafe;
vector[N_cafe] beta_cafe;
real alpha; // Like bar_alpha (hyper-prior)
real beta; // Like bar_beta (hyper-prior)
corr_matrix[N_periods] Rho;
}
model {
vector[N] mu;
//(sigma) - Prior to likelihood
sigma ~ exponential(1);
// (sigma_cafe) - Hyper Prior
sigma_cafe ~ exponential(1);
// (Rho) - Hyper Prior
Rho ~ lkj_corr(2);
// (MU_cafe) - Hyper Prior
alpha ~ normal(5, 2);
beta ~ normal(-1, 0.5);
for (i in 1:N_cafe){
[alpha_cafe[i], beta_cafe[i]]' ~ multi_normal([alpha, beta]', quad_form_diag(Rho, sigma_cafe));
}
// (mu) - Linear model
for (i in 1:N){
mu[i] = alpha_cafe[ cafe[i] ] + beta_cafe[ cafe[i] ] * afternoon[i];
}
// Likelihood
wait ~ normal(mu, sigma);
}
"""
m14_1 = az.from_pystan(
posterior=samples,
posterior_model=posteriori,
observed_data=dat_list.keys()
)
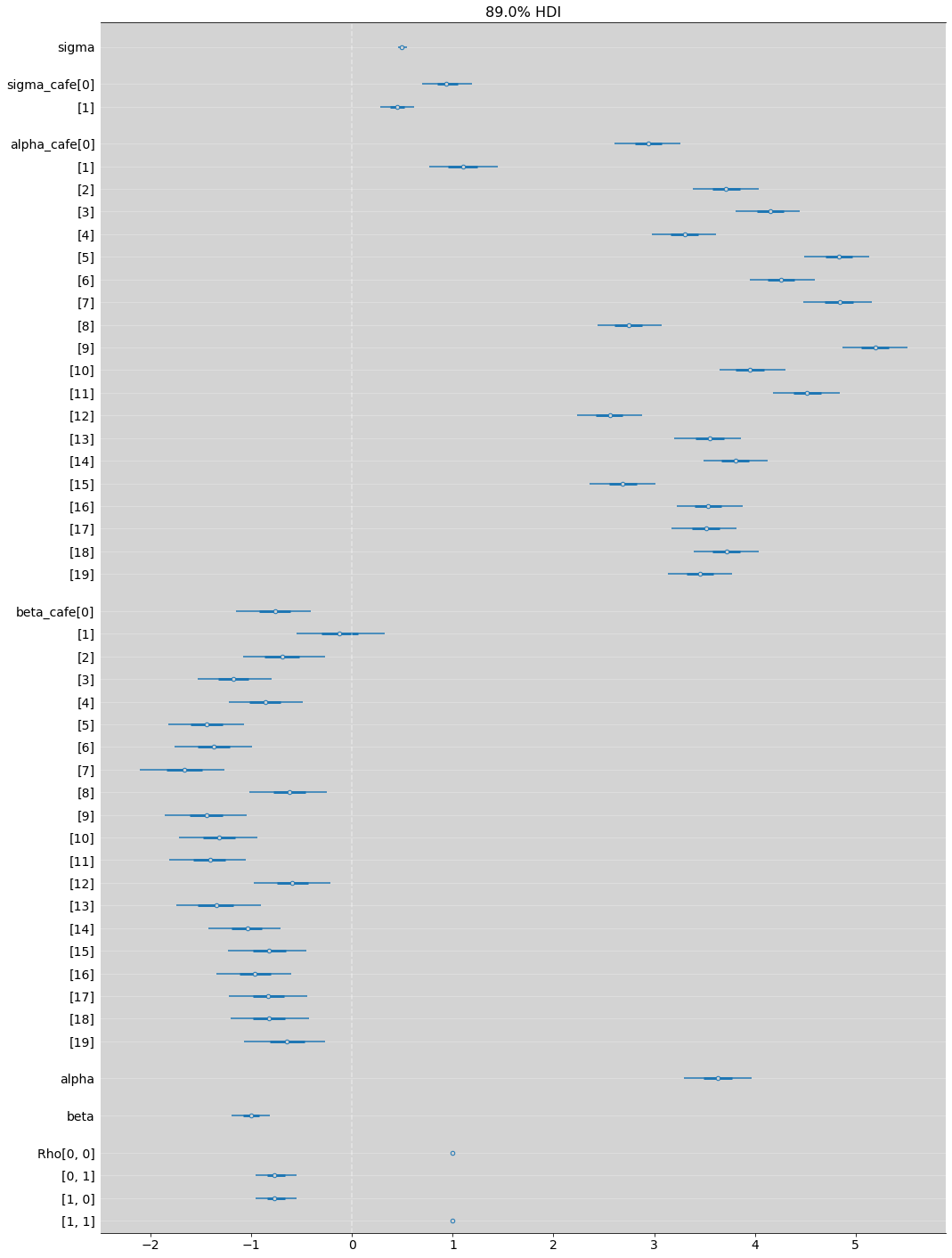
az.summary(m14_1, hdi_prob=0.89)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
| mean | sd | hdi_5.5% | hdi_94.5% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| sigma | 0.494 | 0.027 | 0.454 | 0.541 | 0.000 | 0.000 | 5147.0 | 3085.0 | 1.0 |
| sigma_cafe[0] | 0.955 | 0.163 | 0.695 | 1.187 | 0.002 | 0.002 | 5092.0 | 3241.0 | 1.0 |
| sigma_cafe[1] | 0.454 | 0.107 | 0.281 | 0.616 | 0.002 | 0.002 | 2209.0 | 1995.0 | 1.0 |
| alpha_cafe[0] | 2.944 | 0.204 | 2.608 | 3.259 | 0.003 | 0.002 | 5598.0 | 3371.0 | 1.0 |
| alpha_cafe[1] | 1.102 | 0.213 | 0.766 | 1.445 | 0.003 | 0.002 | 5163.0 | 3691.0 | 1.0 |
| alpha_cafe[2] | 3.714 | 0.208 | 3.384 | 4.043 | 0.003 | 0.002 | 4668.0 | 2735.0 | 1.0 |
| alpha_cafe[3] | 4.153 | 0.199 | 3.807 | 4.447 | 0.003 | 0.002 | 5745.0 | 3062.0 | 1.0 |
| alpha_cafe[4] | 3.306 | 0.203 | 2.976 | 3.615 | 0.003 | 0.002 | 4642.0 | 3251.0 | 1.0 |
| alpha_cafe[5] | 4.836 | 0.202 | 4.486 | 5.135 | 0.003 | 0.002 | 5839.0 | 3544.0 | 1.0 |
| alpha_cafe[6] | 4.259 | 0.203 | 3.948 | 4.598 | 0.003 | 0.002 | 5354.0 | 3489.0 | 1.0 |
| alpha_cafe[7] | 4.835 | 0.215 | 4.478 | 5.165 | 0.003 | 0.002 | 4494.0 | 3171.0 | 1.0 |
| alpha_cafe[8] | 2.744 | 0.206 | 2.436 | 3.075 | 0.003 | 0.002 | 5784.0 | 2714.0 | 1.0 |
| alpha_cafe[9] | 5.194 | 0.204 | 4.869 | 5.515 | 0.003 | 0.002 | 4640.0 | 3116.0 | 1.0 |
| alpha_cafe[10] | 3.951 | 0.205 | 3.654 | 4.303 | 0.003 | 0.002 | 4219.0 | 2878.0 | 1.0 |
| alpha_cafe[11] | 4.524 | 0.208 | 4.181 | 4.844 | 0.003 | 0.002 | 5084.0 | 3117.0 | 1.0 |
| alpha_cafe[12] | 2.557 | 0.202 | 2.230 | 2.881 | 0.003 | 0.002 | 4584.0 | 2660.0 | 1.0 |
| alpha_cafe[13] | 3.554 | 0.210 | 3.198 | 3.864 | 0.003 | 0.002 | 4083.0 | 2590.0 | 1.0 |
| alpha_cafe[14] | 3.806 | 0.202 | 3.487 | 4.124 | 0.003 | 0.002 | 5093.0 | 3223.0 | 1.0 |
| alpha_cafe[15] | 2.686 | 0.206 | 2.354 | 3.012 | 0.003 | 0.002 | 4761.0 | 3236.0 | 1.0 |
| alpha_cafe[16] | 3.537 | 0.202 | 3.228 | 3.880 | 0.003 | 0.002 | 5647.0 | 3117.0 | 1.0 |
| alpha_cafe[17] | 3.514 | 0.204 | 3.171 | 3.821 | 0.003 | 0.002 | 5429.0 | 3417.0 | 1.0 |
| alpha_cafe[18] | 3.718 | 0.205 | 3.395 | 4.041 | 0.003 | 0.002 | 5135.0 | 3168.0 | 1.0 |
| alpha_cafe[19] | 3.453 | 0.202 | 3.135 | 3.776 | 0.003 | 0.002 | 5510.0 | 3590.0 | 1.0 |
| beta_cafe[0] | -0.766 | 0.234 | -1.154 | -0.414 | 0.003 | 0.002 | 5345.0 | 3726.0 | 1.0 |
| beta_cafe[1] | -0.119 | 0.275 | -0.549 | 0.324 | 0.004 | 0.004 | 4223.0 | 2961.0 | 1.0 |
| beta_cafe[2] | -0.686 | 0.259 | -1.080 | -0.271 | 0.004 | 0.003 | 3532.0 | 2484.0 | 1.0 |
| beta_cafe[3] | -1.178 | 0.231 | -1.534 | -0.799 | 0.003 | 0.002 | 5704.0 | 2683.0 | 1.0 |
| beta_cafe[4] | -0.863 | 0.228 | -1.222 | -0.490 | 0.003 | 0.002 | 4366.0 | 3321.0 | 1.0 |
| beta_cafe[5] | -1.450 | 0.238 | -1.821 | -1.070 | 0.003 | 0.002 | 5271.0 | 3484.0 | 1.0 |
| beta_cafe[6] | -1.378 | 0.240 | -1.762 | -0.991 | 0.004 | 0.003 | 4525.0 | 3184.0 | 1.0 |
| beta_cafe[7] | -1.673 | 0.263 | -2.112 | -1.267 | 0.004 | 0.003 | 3798.0 | 3061.0 | 1.0 |
| beta_cafe[8] | -0.619 | 0.243 | -1.024 | -0.248 | 0.003 | 0.003 | 5271.0 | 3084.0 | 1.0 |
| beta_cafe[9] | -1.446 | 0.253 | -1.856 | -1.047 | 0.004 | 0.003 | 4326.0 | 2741.0 | 1.0 |
| beta_cafe[10] | -1.324 | 0.245 | -1.719 | -0.944 | 0.004 | 0.003 | 3760.0 | 2877.0 | 1.0 |
| beta_cafe[11] | -1.421 | 0.243 | -1.819 | -1.054 | 0.004 | 0.003 | 4647.0 | 3288.0 | 1.0 |
| beta_cafe[12] | -0.591 | 0.239 | -0.978 | -0.216 | 0.003 | 0.002 | 4727.0 | 3014.0 | 1.0 |
| beta_cafe[13] | -1.357 | 0.265 | -1.749 | -0.909 | 0.005 | 0.004 | 2824.0 | 2721.0 | 1.0 |
| beta_cafe[14] | -1.045 | 0.229 | -1.428 | -0.709 | 0.003 | 0.002 | 5144.0 | 3521.0 | 1.0 |
| beta_cafe[15] | -0.826 | 0.244 | -1.234 | -0.458 | 0.004 | 0.003 | 4317.0 | 3206.0 | 1.0 |
| beta_cafe[16] | -0.966 | 0.234 | -1.344 | -0.607 | 0.003 | 0.002 | 4929.0 | 3126.0 | 1.0 |
| beta_cafe[17] | -0.826 | 0.241 | -1.220 | -0.445 | 0.003 | 0.002 | 5018.0 | 3167.0 | 1.0 |
| beta_cafe[18] | -0.821 | 0.244 | -1.203 | -0.425 | 0.004 | 0.003 | 4382.0 | 2956.0 | 1.0 |
| beta_cafe[19] | -0.640 | 0.251 | -1.071 | -0.268 | 0.004 | 0.003 | 3712.0 | 3520.0 | 1.0 |
| alpha | 3.632 | 0.216 | 3.293 | 3.972 | 0.003 | 0.002 | 4168.0 | 3264.0 | 1.0 |
| beta | -1.003 | 0.120 | -1.196 | -0.814 | 0.002 | 0.001 | 3412.0 | 3079.0 | 1.0 |
| Rho[0, 0] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 4000.0 | 4000.0 | NaN |
| Rho[0, 1] | -0.743 | 0.142 | -0.961 | -0.553 | 0.003 | 0.002 | 2243.0 | 1976.0 | 1.0 |
| Rho[1, 0] | -0.743 | 0.142 | -0.961 | -0.553 | 0.003 | 0.002 | 2243.0 | 1976.0 | 1.0 |
| Rho[1, 1] | 1.000 | 0.000 | 1.000 | 1.000 | 0.000 | 0.000 | 3969.0 | 3852.0 | 1.0 |
az.plot_forest(m14_1, hdi_prob=0.89, combined=True, figsize=(17, 25))
plt.grid(axis='y', color='white', alpha=0.3)
plt.axvline(x=0, ls='--', color='white', alpha=0.4)
plt.show()
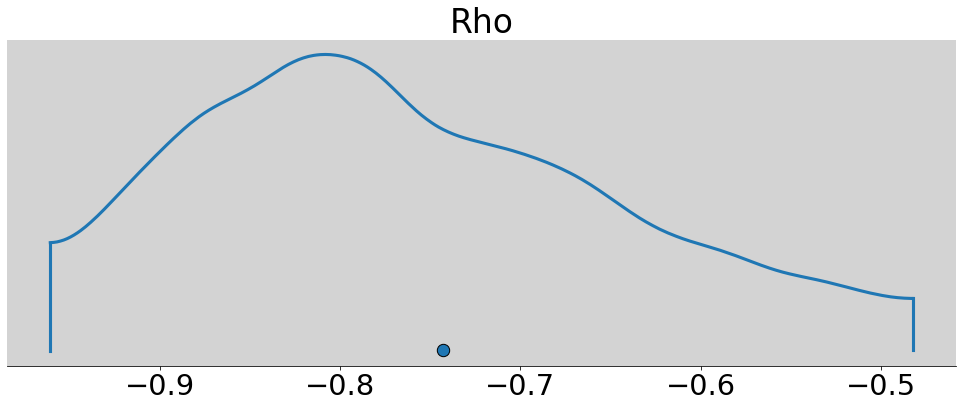
R Code 14.13¶
az.plot_density(m14_1.posterior.Rho.sel(Rho_dim_0=0, Rho_dim_1=1), figsize=(17, 6))
plt.xlabel = 'Correlation'
plt.ylabel = 'Density'
plt.show()
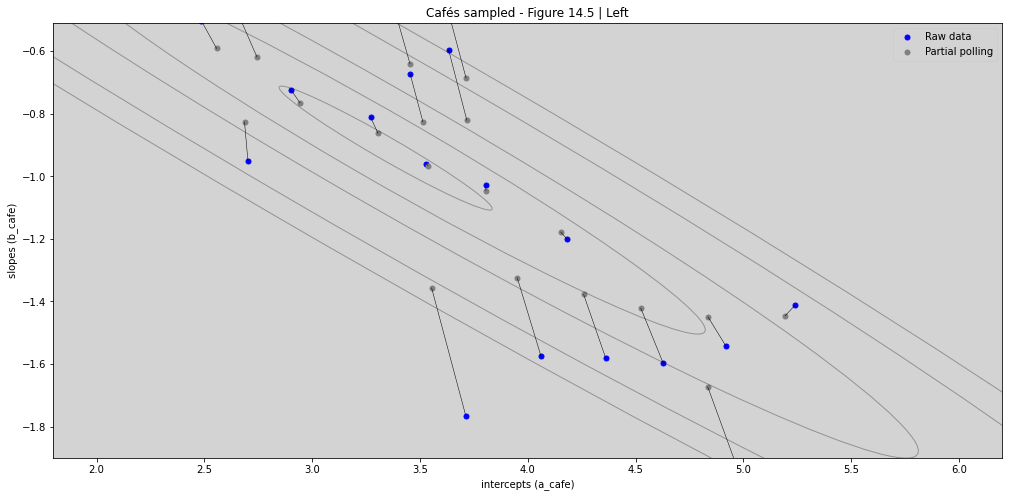
R Code 14.14 & R Code 14.15¶
post_alpha = m14_1.posterior.alpha.mean().values
post_beta = m14_1.posterior.beta.mean().values
post_sigma_alpha = m14_1.posterior.sigma_cafe.sel(sigma_cafe_dim_0=0).mean().values
post_sigma_beta = m14_1.posterior.sigma_cafe.sel(sigma_cafe_dim_0=1).mean().values
post_rho = m14_1.posterior.Rho.sel(Rho_dim_0=0, Rho_dim_1=1).mean().values
print('Posterior alpha:', post_alpha)
print('Posterior beta:', post_beta)
print('Posterior sigma alpha:', post_sigma_alpha)
print('Posterior sigma beta:', post_sigma_beta)
print('Posterior Rho:', post_rho)
Posterior alpha: 3.6320048659560236
Posterior beta: -1.0031288413854578
Posterior sigma alpha: 0.9549508114681846
Posterior sigma beta: 0.4542529928057041
Posterior Rho: -0.7428039489326627
post_matrix_sigma_alpha_beta = np.matrix([[post_sigma_alpha, 0],[0, post_sigma_beta]])
post_matrix_sigma_alpha_beta
matrix([[0.95495081, 0. ],
[0. , 0.45425299]])
post_matrix_rho = np.matrix([[1, post_rho],[post_rho, 1]])
post_matrix_rho
matrix([[ 1. , -0.74280395],
[-0.74280395, 1. ]])
post_Sigma = post_matrix_sigma_alpha_beta * post_matrix_rho * post_matrix_sigma_alpha_beta
post_Sigma
matrix([[ 0.91193105, -0.32222038],
[-0.32222038, 0.20634578]])
a1 = [d.loc[(d['afternoon'] == 0) & (d['cafe'] == i)]['wait'].mean() for i in range(1, N_cafe+1)]
b1 = [d.loc[(d['afternoon'] == 1) & (d['cafe'] == i)]['wait'].mean() for i in range(1, N_cafe+1)]
# Remember that in R Code 14.1 above, the data is build as
# a = 3.5 # Average morning wait time
# b = (-1) # Average difference afternoon wait time
# because this, b value is b - a, like this
b1 = [b1[i] - a1[i] for i in range(len(a1))]
# Extract posterior means of partially polled estimates
a2 = [m14_1.posterior.alpha_cafe.sel(alpha_cafe_dim_0=i).mean().values for i in range(N_cafe)]
b2 = [m14_1.posterior.beta_cafe.sel(beta_cafe_dim_0=i).mean().values for i in range(N_cafe)]
fig, ax_nstd = plt.subplots(figsize=(17, 8))
mu = np.array([post_alpha, post_beta])
dependency_nstd = post_Sigma.tolist()
scale = 5, 5
level = [0.1, 0.3, 0.5, 0.8, 0.99]
x, y = get_correlated_dataset(500, dependency_nstd, mu, scale)
ax_nstd.scatter(a1, b1, s=25, color='b')
ax_nstd.scatter(a2, b2, s=25, facecolor='gray')
ax_nstd.plot([a1, a2], [b1, b2], 'k-', linewidth=0.5)
for l in level:
confidence_ellipse(x, y, ax_nstd, n_std=l, edgecolor='black', alpha=0.3)
# ax_nstd.scatter(mu[0], mu[1], c='red', s=3)
ax_nstd.set_title('Cafés sampled - Figure 14.5 | Left')
ax_nstd.set_xlabel('intercepts (a_cafe)')
ax_nstd.set_ylabel('slopes (b_cafe)')
ax_nstd.set_xlim(1.8, 6.2)
ax_nstd.set_ylim(-1.9, -0.51)
# ax_nstd.legend()
ax_nstd.legend(['Raw data', 'Partial polling'])
plt.plot(1,2, c='r')
plt.show()
R Code 14.16 & R Code 14.17¶
wait_morning_1 = (a1)
wait_afternoon_1 = [sum(x) for x in zip(a1, b1)]
wait_morning_2 = (a2)
wait_afternoon_2 = [sum(x) for x in zip(a2, b2)]
# now shrinkage distribution by simulation
v = np.random.multivariate_normal(mu ,post_Sigma, 1000)
v[:, 1] = [sum(x) for x in v] # calculation afternoon wait
Sigma_est2 = np.cov(v[:, 0], v[:, 1])
mu_est2 = [0, 0] # Empty vector mu
mu_est2[0] = mu[0]
mu_est2[1] = mu[0] + mu[1]
fig, ax_nstd = plt.subplots(figsize=(17, 8))
scale = 5, 5
#level = [0.1, 0.3, 0.5, 0.8, 0.99]
level = [0.1, 0.69, 1.5, 2.0, 3.0]
dependency_nstd = Sigma_est2.tolist()
x, y = get_correlated_dataset(500, dependency_nstd, mu_est2, scale)
ax_nstd.scatter(wait_morning_1, wait_afternoon_1, s=25, color='b')
ax_nstd.scatter(wait_morning_2, wait_afternoon_2, s=25, facecolor='gray')
ax_nstd.plot([wait_morning_1, wait_morning_2],
[wait_afternoon_1, wait_afternoon_2],
'k-', linewidth=0.5)
for l in level:
confidence_ellipse(x, y, ax_nstd, n_std=l, edgecolor='black', alpha=0.3)
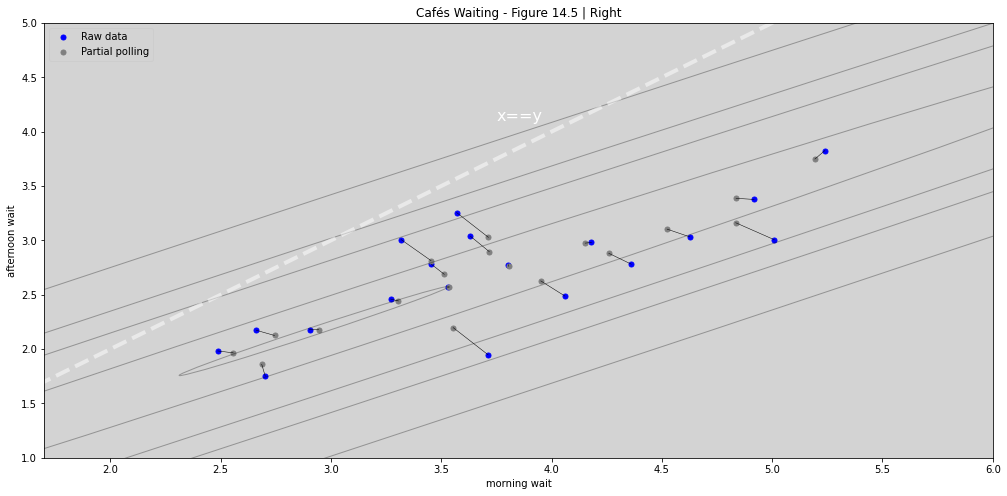
ax_nstd.set_title('Cafés Waiting - Figure 14.5 | Right')
ax_nstd.set_xlabel('morning wait')
ax_nstd.set_ylabel('afternoon wait')
ax_nstd.plot([0, 10], [0, 10], ls='--', color='white', alpha=0.5, linewidth=4)
ax_nstd.text(3.75, 4.1, "x==y",color='white', fontsize=16)
ax_nstd.set_xlim(1.7, 6)
ax_nstd.set_ylim(1, 5)
# ax_nstd.legend()
ax_nstd.legend(['Raw data', 'Partial polling'])
plt.plot(1,2, c='r')
plt.show()
14.2 Advanced varing Slopes¶
R Code 14.18¶
# Previous chimpanzees models is in chapter 11
df = pd.read_csv('./data/chimpanzees.csv', sep=';')
# Build the treatment data
df['treatment'] = 1 + df['prosoc_left'] + 2 * df['condition']
df.head(20)
| actor | recipient | condition | block | trial | prosoc_left | chose_prosoc | pulled_left | treatment | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | NaN | 0 | 1 | 2 | 0 | 1 | 0 | 1 |
| 1 | 1 | NaN | 0 | 1 | 4 | 0 | 0 | 1 | 1 |
| 2 | 1 | NaN | 0 | 1 | 6 | 1 | 0 | 0 | 2 |
| 3 | 1 | NaN | 0 | 1 | 8 | 0 | 1 | 0 | 1 |
| 4 | 1 | NaN | 0 | 1 | 10 | 1 | 1 | 1 | 2 |
| 5 | 1 | NaN | 0 | 1 | 12 | 1 | 1 | 1 | 2 |
| 6 | 1 | NaN | 0 | 2 | 14 | 1 | 0 | 0 | 2 |
| 7 | 1 | NaN | 0 | 2 | 16 | 1 | 0 | 0 | 2 |
| 8 | 1 | NaN | 0 | 2 | 18 | 0 | 1 | 0 | 1 |
| 9 | 1 | NaN | 0 | 2 | 20 | 0 | 1 | 0 | 1 |
| 10 | 1 | NaN | 0 | 2 | 22 | 0 | 0 | 1 | 1 |
| 11 | 1 | NaN | 0 | 2 | 24 | 1 | 0 | 0 | 2 |
| 12 | 1 | NaN | 0 | 3 | 26 | 0 | 0 | 1 | 1 |
| 13 | 1 | NaN | 0 | 3 | 28 | 1 | 1 | 1 | 2 |
| 14 | 1 | NaN | 0 | 3 | 30 | 0 | 1 | 0 | 1 |
| 15 | 1 | NaN | 0 | 3 | 32 | 1 | 1 | 1 | 2 |
| 16 | 1 | NaN | 0 | 3 | 34 | 1 | 0 | 0 | 2 |
| 17 | 1 | NaN | 0 | 3 | 36 | 0 | 1 | 0 | 1 |
| 18 | 1 | NaN | 0 | 4 | 38 | 1 | 1 | 1 | 2 |
| 19 | 1 | NaN | 0 | 4 | 40 | 0 | 0 | 1 | 1 |
df.tail(20)
| actor | recipient | condition | block | trial | prosoc_left | chose_prosoc | pulled_left | treatment | |
|---|---|---|---|---|---|---|---|---|---|
| 484 | 7 | 2.0 | 1 | 3 | 34 | 0 | 0 | 1 | 3 |
| 485 | 7 | 3.0 | 1 | 3 | 36 | 1 | 1 | 1 | 4 |
| 486 | 7 | 7.0 | 1 | 4 | 38 | 0 | 0 | 1 | 3 |
| 487 | 7 | 2.0 | 1 | 4 | 40 | 0 | 0 | 1 | 3 |
| 488 | 7 | 4.0 | 1 | 4 | 42 | 1 | 1 | 1 | 4 |
| 489 | 7 | 5.0 | 1 | 4 | 44 | 0 | 0 | 1 | 3 |
| 490 | 7 | 6.0 | 1 | 4 | 46 | 0 | 0 | 1 | 3 |
| 491 | 7 | 3.0 | 1 | 4 | 48 | 0 | 0 | 1 | 3 |
| 492 | 7 | 5.0 | 1 | 5 | 50 | 1 | 1 | 1 | 4 |
| 493 | 7 | 4.0 | 1 | 5 | 52 | 0 | 0 | 1 | 3 |
| 494 | 7 | 3.0 | 1 | 5 | 54 | 0 | 0 | 1 | 3 |
| 495 | 7 | 2.0 | 1 | 5 | 56 | 1 | 1 | 1 | 4 |
| 496 | 7 | 6.0 | 1 | 5 | 58 | 1 | 1 | 1 | 4 |
| 497 | 7 | 7.0 | 1 | 5 | 60 | 1 | 1 | 1 | 4 |
| 498 | 7 | 5.0 | 1 | 6 | 62 | 1 | 1 | 1 | 4 |
| 499 | 7 | 4.0 | 1 | 6 | 64 | 1 | 1 | 1 | 4 |
| 500 | 7 | 6.0 | 1 | 6 | 66 | 1 | 1 | 1 | 4 |
| 501 | 7 | 3.0 | 1 | 6 | 68 | 0 | 0 | 1 | 3 |
| 502 | 7 | 7.0 | 1 | 6 | 70 | 0 | 0 | 1 | 3 |
| 503 | 7 | 2.0 | 1 | 6 | 72 | 0 | 0 | 1 | 3 |
Hyper-prior to \(S_{_{ACTOR}}\):
Hyper-prior to \(S_{_{BLOCK}}\): $\( \sigma_{_{BLOCK}} \sim Exponential(1) \)$
This is essentially an interaction model that allow the effect of each treatment to vary by actor and each block.
Important features to understanding this model¶
5.1 Overview of data types¶
Array types (stan docs link)¶
array[10] real x;
array[6, 7] matrix[3, 3] m;
array[12, 8, 15] complex z;
declares \(x\) to be a one-dimensional array of size \(10\) containing real values.
declares \(m\) to be a two-dimensional array of size \([6 \times 7]\) containing values that are \([3 \times 3]\) matrices .
declares \(z\) to be a \([12 \times 8 \times 15]\) array of complex numbers.
And read this two parts below
5.5 Vector and matrix data types¶
Stan provides three types of container objects: arrays, vectors, and matrices.
5.6 Array data type¶
Stan supports arrays of arbitrary dimension.
Depreciated Features in Stan to build this example:¶
After Stan 2.26 the declarations below:
int n[5];
real a[3, 4];
real<lower=0> z[5, 4, 2];
vector[7] mu[3];
matrix[7, 2] mu[15, 12];
cholesky_factor_cov[5, 6] mu[2, 3, 4];
was replaced to:
array[5] int n;
array[3, 4] real a;
array[5, 4, 2] real<lower=0> z;
array[3] vector[7] mu;
array[15, 12] matrix[7, 2] mu;
array[2, 3, 4] cholesky_factor_cov[5, 6] mu;
This statements
a[:][1] = b;
was replaced by
a[:, 1] = b;
Explanation:¶
Why this alpha ~ multi_normal(rep_vector(0, 4), quad_form_diag(Rho_actor, sigma_actor)); works in code below?
Response:¶
Let be the code:
parameters{
...
vector[qty_treatments] gamma;
...
}
There are \(4\) positions in vector gamma, then when we atrribute the normal(0, 1), we realize make the broadcast:
gamma ~ normal(0, 1);
is equivalent to:
for (i in 1:qty_treatments){
gamma[i] ~ normal(0, 1);
}
This is the same reason that code with multi_normal works. Let see this in more details:
parameters{
...
array[qty_chimpanzees] vector[qty_treatments] alpha;
...
}
Now, the alpha have two dimensional structure.
Note: index in Stan start in 1, not in zero like python.
The
alpha[1, 1]isalphatochimpanzees=1and totreatment=1The
alpha[1, 2]isalphatochimpanzees=1and totreatment=2The
alpha[2, 3]isalphatochimpanzees=2and totreatment=3The
alpha[7, 4]isalphatochimpanzees=7and totreatment=4
And
The
alpha[1, :]isalphatochimpanzees=1and to alltreatments
The type of data to alpha[1, :] is a vector[qty_treatments].
This is the same type that multi_normal expected to attribuite the results.
Then, the code works!
model = """
data {
int N;
int qty_chimpanzees;
int qty_blocks;
int qty_treatments;
array[N] int pulled_left;
array[N] int actor;
array[N] int block;
array[N] int treatment;
}
parameters{
vector[qty_treatments] gamma;
array[qty_chimpanzees] vector[qty_treatments] alpha;
array[qty_blocks] vector[qty_treatments] beta;
corr_matrix[qty_treatments] Rho_actor;
vector<lower=0>[qty_treatments] sigma_actor;
corr_matrix[qty_treatments] Rho_block;
vector[qty_treatments] sigma_block;
}
model {
vector[N] p;
// Fixed Priors
Rho_actor ~ lkj_corr(4);
sigma_actor ~ exponential(1);
Rho_block ~ lkj_corr(4);
sigma_block ~ exponential(1);
// Adaptatives Priors
gamma ~ normal(0, 1);
alpha ~ multi_normal(rep_vector(0, 4), quad_form_diag(Rho_actor, sigma_actor));
beta ~ multi_normal(rep_vector(0, 4), quad_form_diag(Rho_block, sigma_block));
// Linear model
for (i in 1:N){
p[i] = gamma[ treatment[i] ] + alpha[ actor[i], treatment[i] ] + beta[ block[i], treatment[i] ];
p[i] = inv_logit(p[i]); // Link function
}
// Likelihood
pulled_left ~ binomial(1, p);
}
"""
dat_list = df[['pulled_left', 'actor', 'block', 'treatment']].to_dict('list')
dat_list['N'] = len(df)
dat_list['qty_chimpanzees'] = len(df['actor'].unique())
dat_list['qty_blocks'] = len(df['block'].unique())
dat_list['qty_treatments'] = len(df['treatment'].unique())
posteriori = stan.build(model, data=dat_list)
samples = posteriori.sample(num_chains=4, num_samples=1000)
m_14_2 = az.from_pystan(
posterior=samples,
posterior_model=posteriori,
observed_data=dat_list.keys()
)
print(az.summary(m_14_2).to_string())
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
mean sd hdi_3% hdi_97% mcse_mean mcse_sd ess_bulk ess_tail r_hat
gamma[0] 0.191 0.528 -0.741 1.254 0.034 0.024 229.0 444.0 1.04
gamma[1] 0.669 0.409 -0.141 1.366 0.021 0.015 353.0 800.0 1.02
gamma[2] 0.027 0.588 -1.045 1.192 0.039 0.028 231.0 242.0 1.01
gamma[3] 0.618 0.580 -0.546 1.639 0.034 0.026 288.0 282.0 1.01
alpha[0, 0] -0.860 0.660 -2.257 0.235 0.040 0.030 275.0 302.0 1.03
alpha[0, 1] -0.540 0.481 -1.472 0.334 0.020 0.015 574.0 758.0 1.01
alpha[0, 2] -0.964 0.716 -2.409 0.406 0.040 0.030 329.0 362.0 1.01
alpha[0, 3] -0.537 0.683 -1.698 0.813 0.040 0.034 308.0 435.0 1.01
alpha[1, 0] 3.124 1.366 1.057 5.688 0.080 0.062 346.0 170.0 1.02
alpha[1, 1] 2.046 1.087 0.447 4.116 0.046 0.034 545.0 524.0 1.01
alpha[1, 2] 4.042 1.512 1.278 6.895 0.077 0.055 370.0 346.0 1.01
alpha[1, 3] 3.206 1.562 0.730 5.942 0.081 0.058 335.0 197.0 1.00
alpha[2, 0] -1.119 0.659 -2.436 -0.053 0.038 0.027 305.0 576.0 1.01
alpha[2, 1] -0.290 0.531 -1.222 0.710 0.029 0.021 310.0 280.0 1.01
alpha[2, 2] -1.562 0.764 -2.987 -0.035 0.039 0.027 386.0 651.0 1.01
alpha[2, 3] -1.201 0.684 -2.455 0.016 0.036 0.025 363.0 906.0 1.01
alpha[3, 0] -0.917 0.641 -2.132 0.234 0.028 0.020 533.0 599.0 1.02
alpha[3, 1] -0.585 0.477 -1.479 0.239 0.023 0.017 423.0 655.0 1.01
alpha[3, 2] -1.927 0.809 -3.411 -0.390 0.046 0.032 304.0 366.0 1.00
alpha[3, 3] -0.904 0.691 -2.166 0.379 0.042 0.030 264.0 479.0 1.01
alpha[4, 0] -0.863 0.656 -2.149 0.370 0.040 0.028 280.0 347.0 1.02
alpha[4, 1] -0.403 0.471 -1.318 0.465 0.018 0.014 661.0 918.0 1.01
alpha[4, 2] -0.991 0.760 -2.466 0.387 0.048 0.034 255.0 402.0 1.01
alpha[4, 3] -0.646 0.684 -1.940 0.572 0.040 0.028 298.0 595.0 1.01
alpha[5, 0] 0.882 0.644 -0.373 2.055 0.031 0.022 407.0 675.0 1.02
alpha[5, 1] -0.097 0.494 -0.961 0.854 0.025 0.018 383.0 784.0 1.01
alpha[5, 2] 0.190 0.730 -1.287 1.439 0.049 0.035 229.0 198.0 1.02
alpha[5, 3] -0.107 0.698 -1.398 1.155 0.045 0.032 242.0 154.0 1.02
alpha[6, 0] 1.095 0.690 -0.242 2.356 0.040 0.029 287.0 431.0 1.01
alpha[6, 1] 0.914 0.617 -0.204 2.085 0.057 0.045 141.0 87.0 1.04
alpha[6, 2] 2.632 0.937 1.073 4.521 0.046 0.032 424.0 654.0 1.01
alpha[6, 3] 2.684 1.296 0.514 5.052 0.078 0.058 288.0 343.0 1.01
beta[0, 0] -0.099 0.334 -0.817 0.501 0.013 0.009 581.0 841.0 1.01
beta[0, 1] -0.054 0.327 -0.811 0.536 0.012 0.009 750.0 906.0 1.00
beta[0, 2] -0.065 0.287 -0.618 0.473 0.011 0.008 611.0 811.0 1.02
beta[0, 3] -0.441 0.509 -1.501 0.289 0.029 0.021 346.0 300.0 1.01
beta[1, 0] 0.075 0.349 -0.595 0.763 0.018 0.013 413.0 365.0 1.02
beta[1, 1] -0.148 0.358 -0.854 0.500 0.018 0.013 474.0 547.0 1.01
beta[1, 2] 0.085 0.302 -0.478 0.664 0.012 0.009 451.0 745.0 1.01
beta[1, 3] 0.182 0.375 -0.488 0.855 0.018 0.014 489.0 748.0 1.01
beta[2, 0] 0.356 0.467 -0.228 1.361 0.032 0.023 275.0 281.0 1.01
beta[2, 1] -0.241 0.385 -1.043 0.359 0.024 0.017 317.0 457.0 1.01
beta[2, 2] 0.011 0.312 -0.559 0.599 0.012 0.009 539.0 584.0 1.01
beta[2, 3] 0.166 0.401 -0.502 1.010 0.020 0.014 406.0 658.0 1.01
beta[3, 0] 0.147 0.354 -0.453 0.912 0.020 0.014 329.0 459.0 1.01
beta[3, 1] 0.107 0.355 -0.583 0.837 0.017 0.012 469.0 550.0 1.02
beta[3, 2] -0.113 0.295 -0.758 0.352 0.011 0.008 700.0 798.0 1.01
beta[3, 3] -0.002 0.399 -0.719 0.783 0.018 0.017 589.0 375.0 1.01
beta[4, 0] -0.214 0.358 -0.922 0.409 0.020 0.014 354.0 508.0 1.01
beta[4, 1] 0.020 0.336 -0.566 0.785 0.015 0.011 425.0 705.0 1.01
beta[4, 2] 0.105 0.288 -0.402 0.719 0.010 0.008 714.0 1075.0 1.02
beta[4, 3] -0.012 0.351 -0.662 0.733 0.016 0.013 524.0 212.0 1.01
beta[5, 0] -0.112 0.342 -0.861 0.499 0.018 0.013 382.0 415.0 1.01
beta[5, 1] 0.421 0.487 -0.220 1.359 0.035 0.025 236.0 835.0 1.01
beta[5, 2] -0.052 0.306 -0.671 0.530 0.014 0.010 487.0 825.0 1.01
beta[5, 3] 0.343 0.474 -0.366 1.294 0.023 0.018 493.0 686.0 1.01
Rho_actor[0, 0] 1.000 0.000 1.000 1.000 0.000 0.000 4000.0 4000.0 NaN
Rho_actor[0, 1] 0.327 0.247 -0.085 0.796 0.011 0.008 552.0 854.0 1.01
Rho_actor[0, 2] 0.375 0.234 -0.043 0.785 0.011 0.008 463.0 411.0 1.01
Rho_actor[0, 3] 0.321 0.237 -0.106 0.763 0.010 0.008 535.0 794.0 1.01
Rho_actor[1, 0] 0.327 0.247 -0.085 0.796 0.011 0.008 552.0 854.0 1.01
Rho_actor[1, 1] 1.000 0.000 1.000 1.000 0.000 0.000 1892.0 1217.0 1.00
Rho_actor[1, 2] 0.350 0.245 -0.106 0.770 0.014 0.010 319.0 674.0 1.02
Rho_actor[1, 3] 0.342 0.256 -0.108 0.804 0.015 0.012 302.0 281.0 1.01
Rho_actor[2, 0] 0.375 0.234 -0.043 0.785 0.011 0.008 463.0 411.0 1.01
Rho_actor[2, 1] 0.350 0.245 -0.106 0.770 0.014 0.010 319.0 674.0 1.02
Rho_actor[2, 2] 1.000 0.000 1.000 1.000 0.000 0.000 3280.0 2445.0 1.00
Rho_actor[2, 3] 0.423 0.232 0.000 0.844 0.009 0.007 596.0 792.0 1.01
Rho_actor[3, 0] 0.321 0.237 -0.106 0.763 0.010 0.008 535.0 794.0 1.01
Rho_actor[3, 1] 0.342 0.256 -0.108 0.804 0.015 0.012 302.0 281.0 1.01
Rho_actor[3, 2] 0.423 0.232 0.000 0.844 0.009 0.007 596.0 792.0 1.01
Rho_actor[3, 3] 1.000 0.000 1.000 1.000 0.000 0.000 1644.0 3040.0 1.00
sigma_actor[0] 1.408 0.533 0.621 2.358 0.030 0.021 288.0 255.0 1.03
sigma_actor[1] 0.934 0.413 0.310 1.690 0.017 0.012 565.0 965.0 1.01
sigma_actor[2] 1.881 0.589 0.957 2.952 0.028 0.019 481.0 591.0 1.01
sigma_actor[3] 1.580 0.624 0.643 2.794 0.035 0.025 330.0 336.0 1.01
Rho_block[0, 0] 1.000 0.000 1.000 1.000 0.000 0.000 4000.0 4000.0 NaN
Rho_block[0, 1] -0.040 0.281 -0.545 0.486 0.012 0.008 598.0 1148.0 1.00
Rho_block[0, 2] -0.040 0.310 -0.603 0.530 0.014 0.011 510.0 152.0 1.00
Rho_block[0, 3] 0.021 0.284 -0.500 0.547 0.011 0.008 743.0 1367.0 1.01
Rho_block[1, 0] -0.040 0.281 -0.545 0.486 0.012 0.008 598.0 1148.0 1.00
Rho_block[1, 1] 1.000 0.000 1.000 1.000 0.000 0.000 2940.0 2255.0 1.00
Rho_block[1, 2] -0.044 0.302 -0.541 0.522 0.017 0.012 309.0 402.0 1.01
Rho_block[1, 3] 0.021 0.298 -0.510 0.525 0.015 0.011 415.0 966.0 1.01
Rho_block[2, 0] -0.040 0.310 -0.603 0.530 0.014 0.011 510.0 152.0 1.00
Rho_block[2, 1] -0.044 0.302 -0.541 0.522 0.017 0.012 309.0 402.0 1.01
Rho_block[2, 2] 1.000 0.000 1.000 1.000 0.000 0.000 1818.0 3308.0 1.00
Rho_block[2, 3] 0.023 0.296 -0.547 0.542 0.014 0.010 438.0 680.0 1.02
Rho_block[3, 0] 0.021 0.284 -0.500 0.547 0.011 0.008 743.0 1367.0 1.01
Rho_block[3, 1] 0.021 0.298 -0.510 0.525 0.015 0.011 415.0 966.0 1.01
Rho_block[3, 2] 0.023 0.296 -0.547 0.542 0.014 0.010 438.0 680.0 1.02
Rho_block[3, 3] 1.000 0.000 1.000 1.000 0.000 0.000 2161.0 3213.0 1.00
sigma_block[0] 0.429 0.336 0.011 1.026 0.026 0.019 156.0 237.0 1.03
sigma_block[1] 0.420 0.331 0.006 0.980 0.023 0.017 154.0 158.0 1.02
sigma_block[2] 0.301 0.242 0.004 0.725 0.014 0.010 202.0 388.0 1.03
sigma_block[3] 0.476 0.369 0.009 1.124 0.026 0.018 169.0 138.0 1.02
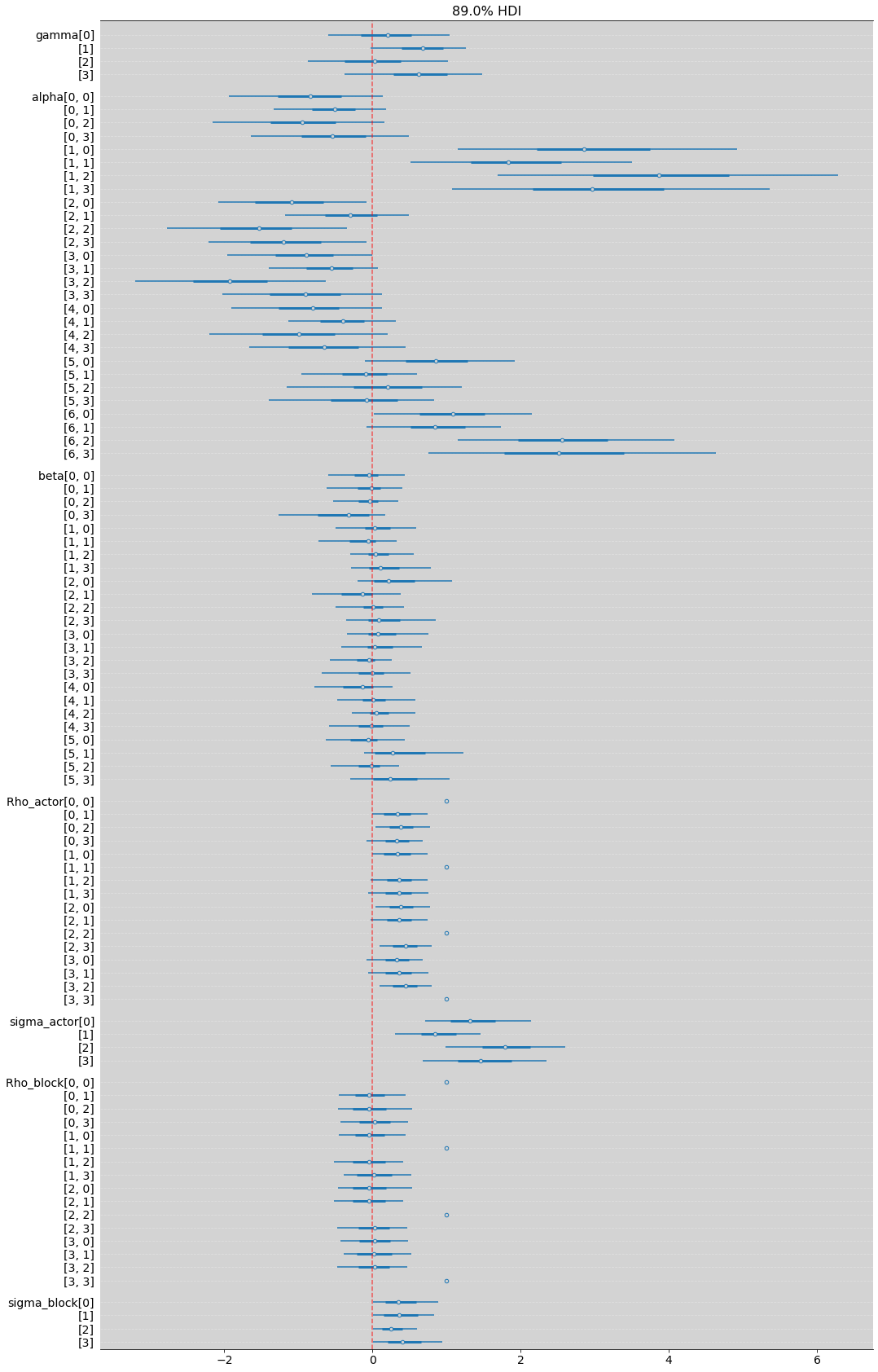
az.plot_forest(m_14_2, hdi_prob=0.89, combined=True, figsize=(17, 30))
plt.grid(axis='y', ls='--', color='white', alpha=0.3)
plt.axvline(x=0, color='red', ls='--', alpha=0.6)
plt.show()
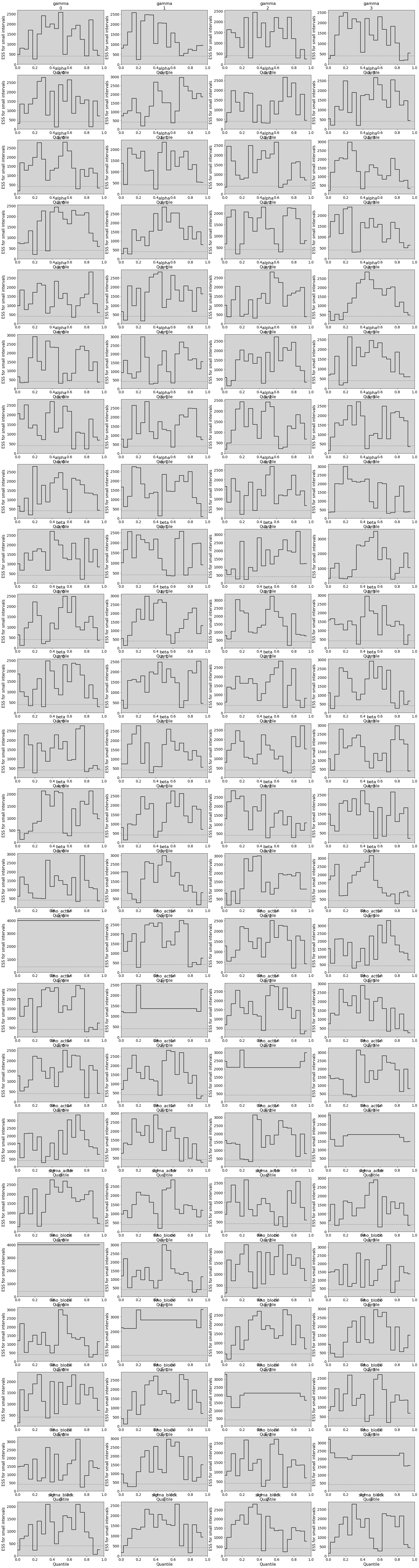
# Like trankplot in this book
az.rcParams['plot.max_subplots'] = 100
az.plot_ess(
m_14_2, kind="local", drawstyle="steps-mid", color="k",
linestyle="-", marker=None
)
plt.show()
R Code 14.19¶
# Undertanding Cholesky
rho_test = 0.3
R_test = np.matrix([[1, rho_test], [rho_test, 1]])
R_test
matrix([[1. , 0.3],
[0.3, 1. ]])
# Cholesky decomposition
cholesky_matrix_L = np.linalg.cholesky(R_test)
cholesky_matrix_L
matrix([[1. , 0. ],
[0.3 , 0.9539392]])
# Testing that R=LL^t
cholesky_matrix_L * np.transpose(cholesky_matrix_L)
matrix([[1. , 0.3],
[0.3, 1. ]])
sample_uncorrelated_z_score = np.random.normal(0, 1, [2,200]) # ?
np.corrcoef(sample_uncorrelated_z_score)
array([[ 1. , -0.15506489],
[-0.15506489, 1. ]])
How this model is works?¶
To understanding the chunck of code below.
transformed parameters{
matrix[qty_treatments, qty_chimpanzees] alpha;
matrix[qty_treatments, qty_blocks] beta;
alpha = (diag_pre_multiply(sigma_actor, L_Rho_actor) * z_actor)';
beta = (diag_pre_multiply(sigma_block, L_Rho_block) * z_block)';
}
PS: \(qty\_actor\) is equal to \(qty\_chimpanzees\) for this example!
Firts, thinking about matrices dimensions:
To z’s (\(z_{anything}\))
z_actor: \([qty\_treatment, qty\_actor]\)z_block: \([qty\_treatment, qty\_block]\)
To sigmas (\(\sigma\)):
sigma_actor: \(\sigma_{_{ACTOR}}[qty\_treatments]\)sigma_block: \(\sigma_{_{BLOCK}}[qty\_treatments]\)
To Rho’s (\(\rho\)):
L_Rho_actor: \(\rho_{_{ACTOR}}[qty\_treatments \times qty\_treatments]\)L_Rho_block: \(\rho_{_{BLOCK}}[qty\_treatments \times qty\_treatments]\)
Now, understanding the stan functions:
The Stan function diag_pre_multiply works:
matrix diag_pre_multiply(vector v, matrix m)Return the product of the diagonal matrix formed from the vector
vand the matrixm, i.e.,diag_matrix(v) * m.
The Stan function diag_matrix(v) works:
matrix diag_matrix(vector x)The diagonal matrix with diagonal x.
For example: \(x = \begin{bmatrix}
0.25 \\
0.30 \\
0.25 \\
0.20
\end{bmatrix}\) then diag_matrix(x) is equal \(\begin{bmatrix}
0.25 & 0 & 0 & 0 \\
0 & 0.30 & 0 & 0 \\
0 & 0 & 0.25 & 0 \\
0 & 0 & 0 & 0.20
\end{bmatrix}\), this is a \([4 \times 4]\) matrix.
Finally, understanding dimensions for alpha and beta:
Then, this diag_pre_multiply(sigma_actor, L_Rho_actor) is:
diag_matrix(sigma_actor)\(\times\)L_Rho_actor\([qty\_treatments \times qty\_treatments] \times [qty\_treatments \times qty\_treatments] = [qty\_treatments \times qty\_treatments]\)
The complete calculations, have this dimension:
(diag_pre_multiply(sigma_actor, L_Rho_actor) * z_actor)\([qty\_treatments \times qty\_treatments] \times [qty\_treatments \times qty\_chimpanzees] = [qty\_treatments \times qty\_chimpanzees]\)
And to finish, transpose the matrix ( ’ ):
(diag_pre_multiply(sigma_actor, L_Rho_actor) * z_actor)'\([qty\_chimpanzees \times qty\_treatments]\)
Each of the index in matrix above, is a prior to alpha (or beta) in the model. Make the model with this structure is not need using the multi_normal(), like the previous model m_14_2.
model = """
data {
int N;
int qty_chimpanzees;
int qty_blocks;
int qty_treatments;
array[N] int pulled_left;
array[N] int actor;
array[N] int block;
array[N] int treatment;
}
parameters{
vector[qty_treatments] gamma;
matrix[qty_treatments, qty_chimpanzees] z_actor;
cholesky_factor_corr[qty_treatments] L_Rho_actor;
vector<lower=0>[qty_treatments] sigma_actor;
matrix[qty_treatments, qty_blocks] z_block;
cholesky_factor_corr[qty_treatments] L_Rho_block;
vector<lower=0>[qty_treatments] sigma_block;
}
transformed parameters{
matrix[qty_chimpanzees, qty_treatments] alpha;
matrix[qty_blocks, qty_treatments] beta;
alpha = (diag_pre_multiply(sigma_actor, L_Rho_actor) * z_actor)'; // matrix[qty_actors, qty_treatments]
beta = (diag_pre_multiply(sigma_block, L_Rho_block) * z_block)'; // Above explanations about this part
}
model {
vector[N] p;
// Adaptative Priors - Non-centered
gamma ~ normal(0, 1);
to_vector(z_actor) ~ normal(0, 1);
L_Rho_actor ~ lkj_corr_cholesky(2);
sigma_actor ~ exponential(1);
to_vector(z_block) ~ normal(0, 1);
L_Rho_block ~ lkj_corr_cholesky(2);
sigma_block ~ exponential(1);
// Linear model
for (i in 1:N){
p[i] = gamma[ treatment[i] ] + alpha[ actor[i], treatment[i] ] + beta[ block[i], treatment[i] ];
p[i] = inv_logit(p[i]); // Link function
}
// Likelihood
pulled_left ~ binomial(1, p);
}
"""
dat_list = df[['pulled_left', 'actor', 'block', 'treatment']].to_dict('list')
dat_list['N'] = len(df)
dat_list['qty_chimpanzees'] = len(df['actor'].unique())
dat_list['qty_blocks'] = len(df['block'].unique())
dat_list['qty_treatments'] = len(df['treatment'].unique())
posteriori = stan.build(model, data=dat_list)
samples = posteriori.sample(num_chains=4, num_samples=1000)
m_14_3 = az.from_pystan(
posterior=samples,
posterior_model=posteriori,
observed_data=dat_list.keys()
)
print(az.summary(m_14_3, hdi_prob=0.89).to_string())
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/diagnostics.py:592: RuntimeWarning: invalid value encountered in double_scalars
(between_chain_variance / within_chain_variance + num_samples - 1) / (num_samples)
mean sd hdi_5.5% hdi_94.5% mcse_mean mcse_sd ess_bulk ess_tail r_hat
gamma[0] 0.214 0.484 -0.542 1.008 0.010 0.007 2269.0 2705.0 1.0
gamma[1] 0.647 0.407 0.032 1.306 0.008 0.006 2707.0 2436.0 1.0
gamma[2] -0.009 0.585 -0.864 0.968 0.010 0.008 3341.0 2985.0 1.0
gamma[3] 0.671 0.548 -0.172 1.536 0.009 0.007 3377.0 2959.0 1.0
z_actor[0, 0] -0.658 0.470 -1.387 0.108 0.009 0.006 2676.0 2969.0 1.0
z_actor[0, 1] 2.352 0.703 1.268 3.497 0.012 0.008 3693.0 3535.0 1.0
z_actor[0, 2] -0.860 0.497 -1.641 -0.076 0.010 0.007 2512.0 2995.0 1.0
z_actor[0, 3] -0.736 0.493 -1.505 0.048 0.010 0.007 2441.0 2824.0 1.0
z_actor[0, 4] -0.643 0.468 -1.395 0.070 0.009 0.006 2762.0 3063.0 1.0
z_actor[0, 5] 0.594 0.481 -0.177 1.337 0.009 0.006 2830.0 3060.0 1.0
z_actor[0, 6] 0.959 0.565 0.054 1.810 0.011 0.008 2879.0 2906.0 1.0
z_actor[1, 0] -0.375 0.667 -1.421 0.704 0.010 0.009 4046.0 3209.0 1.0
z_actor[1, 1] 1.410 0.923 -0.070 2.856 0.015 0.011 3558.0 3111.0 1.0
z_actor[1, 2] -0.040 0.679 -1.112 1.050 0.012 0.009 3032.0 2925.0 1.0
z_actor[1, 3] -0.438 0.688 -1.586 0.616 0.012 0.009 3536.0 3026.0 1.0
z_actor[1, 4] -0.200 0.653 -1.248 0.814 0.011 0.009 3465.0 3298.0 1.0
z_actor[1, 5] -0.360 0.664 -1.430 0.663 0.010 0.009 4207.0 3114.0 1.0
z_actor[1, 6] 0.774 0.799 -0.428 2.097 0.014 0.010 3292.0 3337.0 1.0
z_actor[2, 0] -0.095 0.658 -1.202 0.861 0.010 0.011 4251.0 3045.0 1.0
z_actor[2, 1] 0.854 0.954 -0.696 2.299 0.014 0.011 4868.0 3715.0 1.0
z_actor[2, 2] -0.512 0.685 -1.579 0.576 0.011 0.009 3935.0 2990.0 1.0
z_actor[2, 3] -0.650 0.685 -1.766 0.410 0.011 0.008 3896.0 3334.0 1.0
z_actor[2, 4] -0.172 0.652 -1.186 0.869 0.010 0.009 4222.0 2968.0 1.0
z_actor[2, 5] -0.102 0.669 -1.088 0.987 0.011 0.009 3832.0 3130.0 1.0
z_actor[2, 6] 0.988 0.853 -0.343 2.307 0.014 0.010 3600.0 3031.0 1.0
z_actor[3, 0] 0.071 0.750 -1.127 1.247 0.010 0.011 5689.0 2879.0 1.0
z_actor[3, 1] 0.466 0.929 -1.026 1.906 0.011 0.013 7144.0 3451.0 1.0
z_actor[3, 2] -0.412 0.766 -1.645 0.780 0.011 0.010 5330.0 3191.0 1.0
z_actor[3, 3] 0.015 0.768 -1.291 1.159 0.010 0.013 5528.0 2981.0 1.0
z_actor[3, 4] -0.088 0.715 -1.262 0.998 0.010 0.011 5285.0 3284.0 1.0
z_actor[3, 5] -0.322 0.753 -1.551 0.835 0.010 0.010 5436.0 3206.0 1.0
z_actor[3, 6] 0.831 0.928 -0.635 2.293 0.011 0.011 6855.0 2990.0 1.0
L_Rho_actor[0, 0] 1.000 0.000 1.000 1.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[0, 1] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[0, 2] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[0, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[1, 0] 0.431 0.276 0.007 0.855 0.005 0.004 2884.0 3014.0 1.0
L_Rho_actor[1, 1] 0.848 0.141 0.650 1.000 0.003 0.002 2922.0 3014.0 1.0
L_Rho_actor[1, 2] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[1, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[2, 0] 0.532 0.248 0.164 0.903 0.004 0.003 3145.0 3045.0 1.0
L_Rho_actor[2, 1] 0.282 0.307 -0.181 0.784 0.006 0.005 2295.0 3168.0 1.0
L_Rho_actor[2, 2] 0.670 0.179 0.398 0.961 0.003 0.002 2869.0 3066.0 1.0
L_Rho_actor[2, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_actor[3, 0] 0.452 0.262 0.050 0.849 0.005 0.004 2657.0 3037.0 1.0
L_Rho_actor[3, 1] 0.274 0.315 -0.185 0.806 0.006 0.004 2731.0 3051.0 1.0
L_Rho_actor[3, 2] 0.293 0.317 -0.204 0.790 0.006 0.004 2498.0 2774.0 1.0
L_Rho_actor[3, 3] 0.579 0.180 0.291 0.865 0.003 0.002 2690.0 3154.0 1.0
sigma_actor[0] 1.383 0.489 0.664 2.025 0.010 0.007 2353.0 3000.0 1.0
sigma_actor[1] 0.901 0.404 0.296 1.434 0.007 0.005 2969.0 2671.0 1.0
sigma_actor[2] 1.845 0.592 0.931 2.608 0.011 0.009 3250.0 2401.0 1.0
sigma_actor[3] 1.568 0.582 0.669 2.312 0.011 0.008 3144.0 2650.0 1.0
z_block[0, 0] -0.279 0.844 -1.581 1.139 0.012 0.012 5021.0 2942.0 1.0
z_block[0, 1] 0.115 0.834 -1.279 1.396 0.011 0.013 5926.0 3115.0 1.0
z_block[0, 2] 0.664 0.865 -0.682 2.039 0.012 0.010 4964.0 3108.0 1.0
z_block[0, 3] 0.268 0.840 -1.125 1.535 0.011 0.014 5965.0 3090.0 1.0
z_block[0, 4] -0.429 0.844 -1.837 0.803 0.012 0.012 5334.0 3088.0 1.0
z_block[0, 5] -0.256 0.885 -1.622 1.198 0.012 0.013 5104.0 3190.0 1.0
z_block[1, 0] -0.139 0.877 -1.574 1.206 0.012 0.014 5667.0 3063.0 1.0
z_block[1, 1] -0.252 0.820 -1.567 1.065 0.010 0.014 6141.0 2950.0 1.0
z_block[1, 2] -0.402 0.841 -1.747 0.953 0.011 0.012 5565.0 3300.0 1.0
z_block[1, 3] 0.240 0.853 -1.050 1.629 0.011 0.013 6536.0 3148.0 1.0
z_block[1, 4] 0.035 0.803 -1.282 1.268 0.010 0.012 6050.0 3079.0 1.0
z_block[1, 5] 0.795 0.882 -0.606 2.209 0.013 0.011 4431.0 3202.0 1.0
z_block[2, 0] -0.128 0.942 -1.655 1.339 0.012 0.015 5812.0 3369.0 1.0
z_block[2, 1] 0.174 0.962 -1.432 1.656 0.011 0.016 7222.0 3020.0 1.0
z_block[2, 2] 0.019 0.938 -1.537 1.437 0.011 0.018 7040.0 2325.0 1.0
z_block[2, 3] -0.268 0.893 -1.619 1.242 0.011 0.014 7031.0 3159.0 1.0
z_block[2, 4] 0.235 0.932 -1.228 1.746 0.011 0.015 7393.0 3028.0 1.0
z_block[2, 5] -0.069 0.926 -1.558 1.387 0.012 0.016 6284.0 3006.0 1.0
z_block[3, 0] -0.614 0.961 -2.149 0.915 0.012 0.012 6496.0 3194.0 1.0
z_block[3, 1] 0.258 0.907 -1.227 1.666 0.011 0.015 6812.0 2742.0 1.0
z_block[3, 2] 0.239 0.900 -1.211 1.650 0.011 0.014 6636.0 3177.0 1.0
z_block[3, 3] -0.012 0.933 -1.528 1.462 0.011 0.016 6781.0 2974.0 1.0
z_block[3, 4] -0.061 0.913 -1.435 1.453 0.011 0.016 7351.0 3078.0 1.0
z_block[3, 5] 0.454 0.930 -1.027 1.931 0.011 0.014 6617.0 2932.0 1.0
L_Rho_block[0, 0] 1.000 0.000 1.000 1.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[0, 1] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[0, 2] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[0, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[1, 0] -0.062 0.374 -0.623 0.574 0.005 0.006 4592.0 3122.0 1.0
L_Rho_block[1, 1] 0.920 0.097 0.794 1.000 0.002 0.001 2692.0 3089.0 1.0
L_Rho_block[1, 2] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[1, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[2, 0] -0.020 0.378 -0.649 0.557 0.004 0.007 7142.0 2844.0 1.0
L_Rho_block[2, 1] -0.035 0.365 -0.602 0.562 0.004 0.006 7624.0 3040.0 1.0
L_Rho_block[2, 2] 0.839 0.135 0.655 1.000 0.003 0.002 1757.0 2150.0 1.0
L_Rho_block[2, 3] 0.000 0.000 0.000 0.000 0.000 0.000 4000.0 4000.0 NaN
L_Rho_block[3, 0] 0.045 0.379 -0.552 0.663 0.005 0.006 5021.0 3052.0 1.0
L_Rho_block[3, 1] 0.056 0.373 -0.487 0.709 0.005 0.006 4887.0 3093.0 1.0
L_Rho_block[3, 2] 0.012 0.373 -0.572 0.627 0.005 0.006 4738.0 2872.0 1.0
L_Rho_block[3, 3] 0.739 0.166 0.508 0.986 0.004 0.003 2069.0 2228.0 1.0
sigma_block[0] 0.400 0.321 0.000 0.796 0.006 0.004 2252.0 2283.0 1.0
sigma_block[1] 0.445 0.344 0.000 0.892 0.007 0.005 1967.0 2353.0 1.0
sigma_block[2] 0.303 0.268 0.000 0.627 0.004 0.003 3314.0 2291.0 1.0
sigma_block[3] 0.473 0.375 0.000 0.938 0.007 0.005 1971.0 2057.0 1.0
alpha[0, 0] -0.844 0.609 -1.798 0.138 0.011 0.008 3018.0 3108.0 1.0
alpha[0, 1] -0.511 0.488 -1.332 0.210 0.008 0.006 3697.0 3196.0 1.0
alpha[0, 2] -0.968 0.724 -2.073 0.196 0.012 0.009 3837.0 3287.0 1.0
alpha[0, 3] -0.573 0.659 -1.688 0.384 0.011 0.008 3787.0 3380.0 1.0
alpha[1, 0] 3.152 1.250 1.301 4.881 0.020 0.015 4111.0 3012.0 1.0
alpha[1, 1] 2.005 0.994 0.487 3.404 0.017 0.012 3564.0 2907.0 1.0
alpha[1, 2] 4.309 1.662 1.993 6.688 0.026 0.019 4333.0 3691.0 1.0
alpha[1, 3] 3.406 1.531 1.284 5.719 0.024 0.018 4224.0 3729.0 1.0
alpha[2, 0] -1.107 0.627 -2.048 -0.059 0.012 0.008 2984.0 2985.0 1.0
alpha[2, 1] -0.326 0.475 -1.061 0.424 0.008 0.006 3701.0 3368.0 1.0
alpha[2, 2] -1.537 0.758 -2.752 -0.397 0.012 0.009 3838.0 3581.0 1.0
alpha[2, 3] -1.298 0.660 -2.315 -0.265 0.011 0.008 3689.0 3640.0 1.0
alpha[3, 0] -0.937 0.609 -1.988 -0.034 0.011 0.008 2847.0 2833.0 1.0
alpha[3, 1] -0.580 0.489 -1.425 0.103 0.008 0.006 3921.0 3536.0 1.0
alpha[3, 2] -1.833 0.810 -3.070 -0.571 0.013 0.009 3685.0 3422.0 1.0
alpha[3, 3] -0.976 0.655 -1.992 0.053 0.010 0.007 3970.0 3603.0 1.0
alpha[4, 0] -0.830 0.606 -1.803 0.092 0.011 0.008 2947.0 3191.0 1.0
alpha[4, 1] -0.376 0.476 -1.180 0.332 0.008 0.006 3636.0 3293.0 1.0
alpha[4, 2] -0.970 0.726 -2.106 0.157 0.012 0.009 3644.0 3421.0 1.0
alpha[4, 3] -0.701 0.649 -1.760 0.301 0.010 0.008 4094.0 3235.0 1.0
alpha[5, 0] 0.781 0.638 -0.226 1.775 0.013 0.009 2588.0 3109.0 1.0
alpha[5, 1] -0.047 0.489 -0.838 0.713 0.008 0.006 3884.0 2572.0 1.0
alpha[5, 2] 0.247 0.705 -0.802 1.415 0.012 0.009 3467.0 3059.0 1.0
alpha[5, 3] -0.099 0.654 -1.140 0.903 0.011 0.008 3396.0 3452.0 1.0
alpha[6, 0] 1.206 0.661 0.164 2.255 0.012 0.009 2952.0 3040.0 1.0
alpha[6, 1] 0.923 0.590 -0.008 1.821 0.010 0.007 3706.0 3387.0 1.0
alpha[6, 2] 2.644 0.948 1.114 4.069 0.015 0.010 4093.0 3203.0 1.0
alpha[6, 3] 2.583 1.238 0.819 4.327 0.021 0.015 3962.0 2851.0 1.0
beta[0, 0] -0.128 0.347 -0.694 0.366 0.005 0.005 4443.0 3501.0 1.0
beta[0, 1] -0.052 0.356 -0.625 0.522 0.005 0.004 5006.0 3834.0 1.0
beta[0, 2] -0.051 0.296 -0.509 0.410 0.005 0.004 3767.0 3333.0 1.0
beta[0, 3] -0.410 0.502 -1.230 0.177 0.009 0.006 2840.0 3496.0 1.0
beta[1, 0] 0.055 0.337 -0.446 0.622 0.005 0.005 4781.0 3152.0 1.0
beta[1, 1] -0.133 0.354 -0.723 0.371 0.005 0.004 4166.0 3478.0 1.0
beta[1, 2] 0.083 0.319 -0.308 0.653 0.005 0.004 4656.0 3749.0 1.0
beta[1, 3] 0.170 0.382 -0.395 0.780 0.006 0.005 3841.0 3438.0 1.0
beta[2, 0] 0.317 0.425 -0.208 0.992 0.007 0.005 3416.0 3690.0 1.0
beta[2, 1] -0.234 0.384 -0.862 0.274 0.006 0.004 4393.0 3765.0 1.0
beta[2, 2] 0.005 0.301 -0.433 0.478 0.005 0.004 4467.0 3665.0 1.0
beta[2, 3] 0.160 0.395 -0.419 0.786 0.007 0.005 3436.0 3374.0 1.0
beta[3, 0] 0.132 0.363 -0.347 0.765 0.006 0.005 4384.0 3346.0 1.0
beta[3, 1] 0.121 0.373 -0.460 0.688 0.006 0.005 4625.0 3475.0 1.0
beta[3, 2] -0.118 0.299 -0.600 0.284 0.005 0.004 3625.0 3715.0 1.0
beta[3, 3] 0.020 0.397 -0.618 0.661 0.006 0.005 4497.0 3784.0 1.0
beta[4, 0] -0.206 0.380 -0.852 0.282 0.006 0.005 3851.0 3106.0 1.0
beta[4, 1] 0.049 0.350 -0.496 0.606 0.005 0.004 4743.0 3521.0 1.0
beta[4, 2] 0.111 0.315 -0.276 0.632 0.005 0.004 4500.0 3759.0 1.0
beta[4, 3] -0.027 0.363 -0.617 0.522 0.006 0.005 4160.0 3394.0 1.0
beta[5, 0] -0.129 0.364 -0.757 0.365 0.005 0.005 4804.0 3337.0 1.0
beta[5, 1] 0.436 0.499 -0.158 1.206 0.010 0.007 2438.0 3590.0 1.0
beta[5, 2] -0.047 0.299 -0.479 0.404 0.005 0.003 3962.0 3423.0 1.0
beta[5, 3] 0.328 0.482 -0.272 1.100 0.008 0.006 3374.0 3766.0 1.0
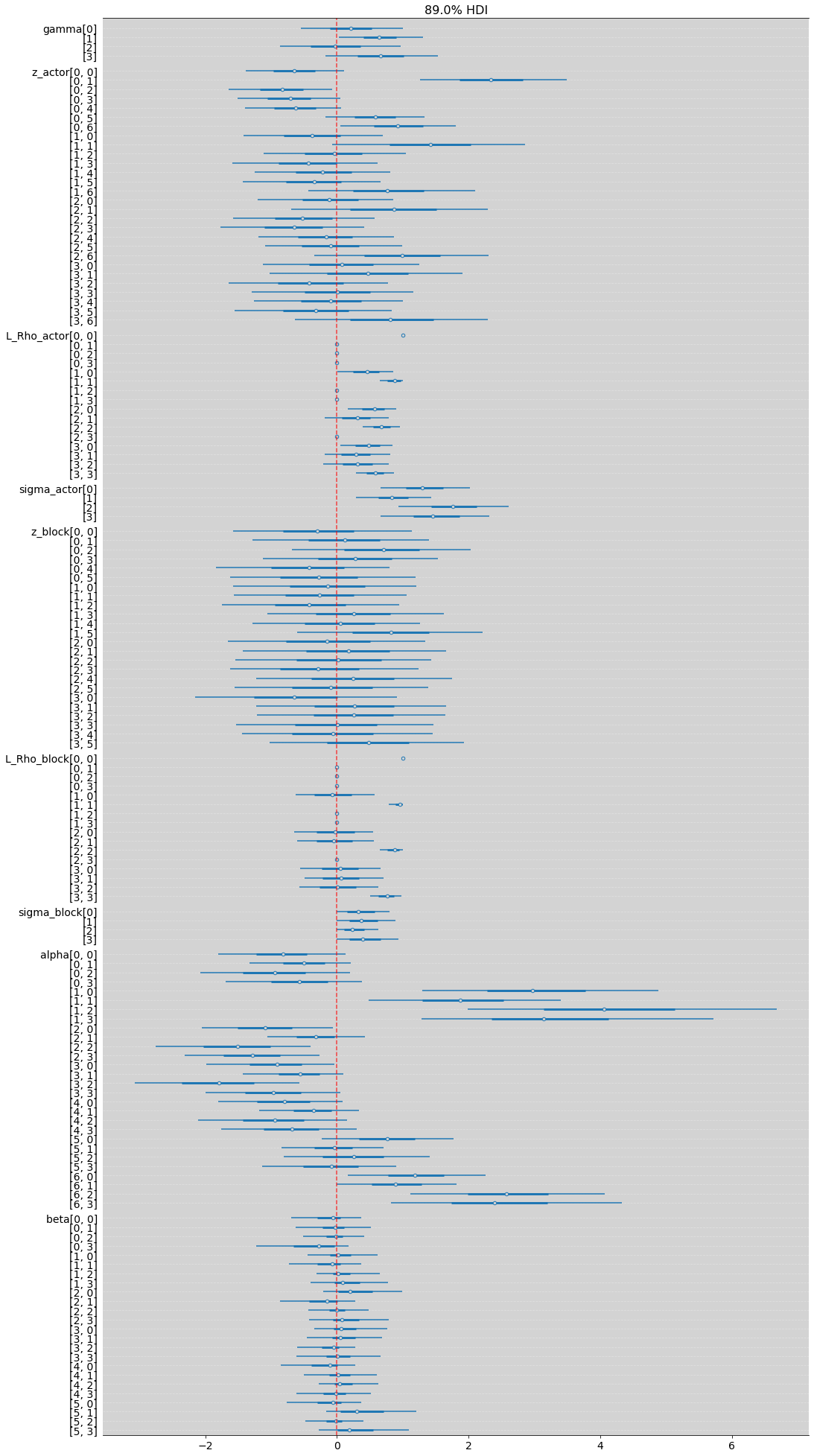
az.plot_forest(m_14_3, combined=True, hdi_prob=0.89, figsize=(17, 35))
plt.axvline(x=0, color='red', ls='--', alpha=0.7)
plt.grid(axis='y', color='white', ls='--', alpha=0.3)
plt.show()
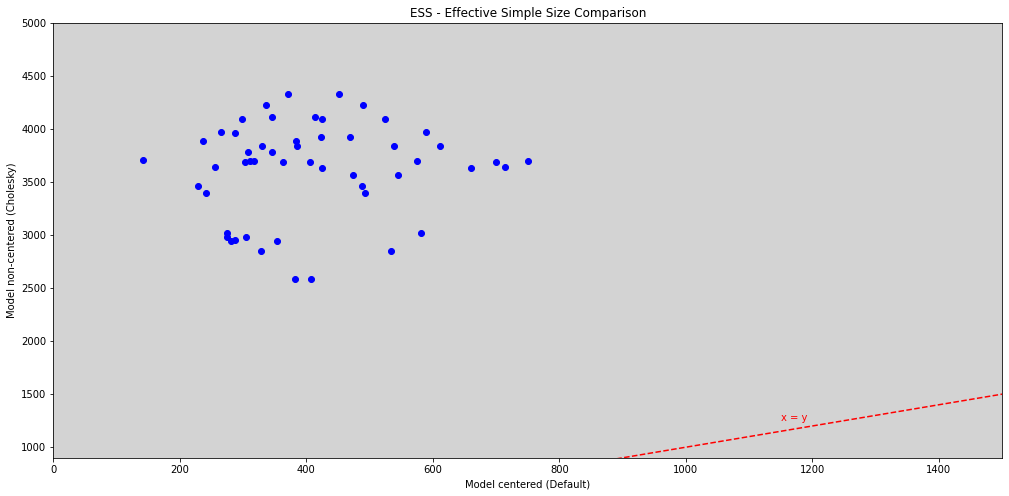
R Code 14.20¶
import matplotlib.pyplot as plt
from importlib import reload
plt=reload(plt)
alpha_ess_14_3 = az.ess(m_14_3, var_names=["alpha", "beta"]).alpha.values.flatten()
beta_ess_14_3 = az.ess(m_14_3, var_names=["alpha", "beta"]).alpha.values.flatten()
alpha_ess_14_2 = az.ess(m_14_2, var_names=["alpha", "beta"]).alpha.values.flatten()
beta_ess_14_2 = az.ess(m_14_2, var_names=["alpha", "beta"]).beta.values.flatten()
plt.figure(figsize=(17, 8))
plt.scatter(alpha_ess_14_2, alpha_ess_14_3, c='b')
plt.scatter(beta_ess_14_2, beta_ess_14_3[:24], c='b')
plt.plot([0, 5000], [0, 5000], ls='--', c='red')
plt.text(1150, 1250, 'x = y', color='red')
plt.ylim(900, 5000)
plt.xlim(0, 1500)
plt.title('ESS - Effective Simple Size Comparison')
plt.xlabel('Model centered (Default)')
plt.ylabel('Model non-centered (Cholesky)')
plt.show()
R Code 14.21¶
az.summary(m_14_3, var_names=['sigma_actor', 'sigma_block'], hdi_prob=0.89)
| mean | sd | hdi_5.5% | hdi_94.5% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| sigma_actor[0] | 1.383 | 0.489 | 0.664 | 2.025 | 0.010 | 0.007 | 2353.0 | 3000.0 | 1.0 |
| sigma_actor[1] | 0.901 | 0.404 | 0.296 | 1.434 | 0.007 | 0.005 | 2969.0 | 2671.0 | 1.0 |
| sigma_actor[2] | 1.845 | 0.592 | 0.931 | 2.608 | 0.011 | 0.009 | 3250.0 | 2401.0 | 1.0 |
| sigma_actor[3] | 1.568 | 0.582 | 0.669 | 2.312 | 0.011 | 0.008 | 3144.0 | 2650.0 | 1.0 |
| sigma_block[0] | 0.400 | 0.321 | 0.000 | 0.796 | 0.006 | 0.004 | 2252.0 | 2283.0 | 1.0 |
| sigma_block[1] | 0.445 | 0.344 | 0.000 | 0.892 | 0.007 | 0.005 | 1967.0 | 2353.0 | 1.0 |
| sigma_block[2] | 0.303 | 0.268 | 0.000 | 0.627 | 0.004 | 0.003 | 3314.0 | 2291.0 | 1.0 |
| sigma_block[3] | 0.473 | 0.375 | 0.000 | 0.938 | 0.007 | 0.005 | 1971.0 | 2057.0 | 1.0 |
R Code 14.22¶
# Like R Code 11.17
# ==================
TODO = """
plt.figure(figsize=(18, 6))
plt.ylim(0, 1.3)
plt.xlim(0, 7)
plt.axhline(y=1, ls='-', c='black')
plt.axhline(y=0.5, ls='--', c='black')
for i in range(7):
plt.axvline(x=i+1, c='black')
plt.text(x=i + 0.25, y=1.1, s=f'Actor {i + 1}', size=20)
alpha_chimp = az.extract(samples_chimpanzees.posterior.alpha.sel(alpha_dim_0=i)).alpha.values
RN = inv_logit(alpha_chimp + az.extract(samples_chimpanzees.posterior.beta.sel(beta_dim_0=0)).beta.values)
LN = inv_logit(alpha_chimp + az.extract(samples_chimpanzees.posterior.beta.sel(beta_dim_0=1)).beta.values)
RP = inv_logit(alpha_chimp + az.extract(samples_chimpanzees.posterior.beta.sel(beta_dim_0=2)).beta.values)
LP = inv_logit(alpha_chimp + az.extract(samples_chimpanzees.posterior.beta.sel(beta_dim_0=3)).beta.values)
# To R/N and R/P
# ===============
if not i == 1:
plt.plot([0.2 + i, 0.8 + i], [RN.mean(), RP.mean()], color='black')
# Plot hdi compatibility interval
RN_hdi_min, RN_hdi_max = az.hdi(RN, hdi_prob=0.89)
RP_hdi_min, RP_hdi_max = az.hdi(RP, hdi_prob=0.89)
plt.plot([0.2 + i, 0.2 + i], [RN_hdi_min, RN_hdi_max], c='black')
plt.plot([0.8 + i, 0.8 + i], [RP_hdi_min, RP_hdi_max], c='black')
# Plot points
plt.plot(0.2 + i, RN.mean(), 'o', markerfacecolor='white', color='black')
plt.plot(0.8 + i, RP.mean(), 'o', markerfacecolor='black', color='white')
# To L/N and L/P
# ===============
if not i == 1:
plt.plot([0.3 + i, 0.9 + i], [LN.mean(), LP.mean()], color='black', linewidth=3)
# Plot hdi compatibility interval
LN_hdi_min, LN_hdi_max = az.hdi(LN, hdi_prob=0.89)
LP_hdi_min, LP_hdi_max = az.hdi(LP, hdi_prob=0.89)
plt.plot([0.3 + i, 0.3 + i], [LN_hdi_min, LN_hdi_max], c='black')
plt.plot([0.9 + i, 0.9 + i], [LP_hdi_min, LP_hdi_max], c='black')
plt.plot(0.3 + i, LN.mean(), 'o', markerfacecolor='white', color='black')
plt.plot(0.9 + i, LP.mean(), 'o', markerfacecolor='black', color='white')
# Labels for only first points
if i == 0:
plt.text(x=0.08, y=RN.mean() - 0.08, s='R/N')
plt.text(x=0.68, y=RP.mean() - 0.08, s='R/P')
plt.text(x=0.17, y=LN.mean() + 0.08, s='L/N')
plt.text(x=0.77, y=LP.mean() + 0.08, s='L/P')
plt.title('Posterior Predictions')
plt.ylabel('Proportion left lever')
plt.yticks([0, 0.5, 1], [0, 0.5, 1])
plt.xticks([])
plt.show()
"""
# TODO: Rebuild to this example.
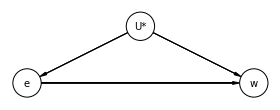
14.3 Instrumental and Causal Designs¶
The variable \(^*U\) is an unobserved.
dag_14_3_1 = CausalGraphicalModel(
nodes=['e', 'U*', 'w'],
edges=[
('U*', 'e'),
('U*', 'w'),
('e', 'w')
],
)
# Drawing the DAG
pgm = daft.PGM()
coordinates = {
"U*": (2, 1),
"e": (0, 0),
"w": (4, 0),
}
for node in dag_14_3_1.dag.nodes:
pgm.add_node(node, node, *coordinates[node])
for edge in dag_14_3_1.dag.edges:
pgm.add_edge(*edge)
pgm.render()
plt.show()
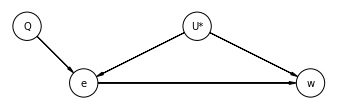
Now, we add Instrumental Variable Q:
dag_14_3_1 = CausalGraphicalModel(
nodes=['e', 'U*', 'w', 'Q'],
edges=[
('U*', 'e'),
('U*', 'w'),
('e', 'w'),
('Q', 'e')
],
)
# Drawing the DAG
pgm = daft.PGM()
coordinates = {
"U*": (3, 1),
"e": (1, 0),
"w": (5, 0),
"Q": (0, 1),
}
for node in dag_14_3_1.dag.nodes:
pgm.add_node(node, node, *coordinates[node])
for edge in dag_14_3_1.dag.edges:
pgm.add_edge(*edge)
pgm.render()
plt.show()
R Code 14.23¶
def standartize(data, get_parameters=False):
mean = np.mean(data)
std = np.std(data)
if not get_parameters:
return list((data - mean)/std)
else:
return list((data - mean)/std), mean, std
N = 500
U_sim = np.random.normal(0, 1, N)
Q_sim = np.random.choice([1,2,3,4], size=N, replace=True)
E_sim = np.random.normal(U_sim + Q_sim, 1, N) # E is influencied by U and Q
W_sim = np.random.normal(U_sim + 0*E_sim, 1, N) # W is influencied only by U, not by E
dat_sim = pd.DataFrame({
'W': standartize(W_sim),
'E': standartize(E_sim),
'Q': standartize(Q_sim)
})
dat_sim
| W | E | Q | |
|---|---|---|---|
| 0 | 0.458919 | -1.050410 | -1.412495 |
| 1 | 0.367655 | 0.827180 | 1.390074 |
| 2 | -0.058669 | 0.095503 | 0.455884 |
| 3 | 0.139368 | 1.715983 | 0.455884 |
| 4 | 1.289500 | 1.171285 | 0.455884 |
| ... | ... | ... | ... |
| 495 | -1.661304 | 0.222796 | 0.455884 |
| 496 | 0.078534 | 0.363780 | 0.455884 |
| 497 | -0.341889 | -1.104266 | -1.412495 |
| 498 | 0.326049 | -1.023097 | -0.478305 |
| 499 | 2.156961 | 2.760130 | 1.390074 |
500 rows × 3 columns
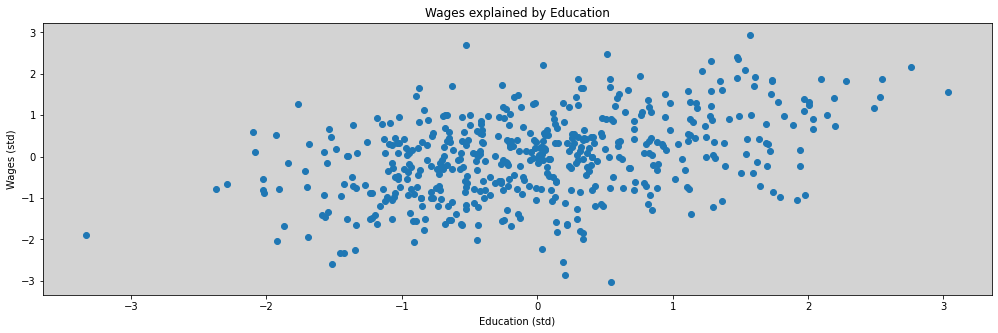
plt.figure(figsize=(17,5))
plt.plot(dat_sim.E, dat_sim.W, 'o')
plt.title('Wages explained by Education')
plt.xlabel('Education (std)')
plt.ylabel('Wages (std)')
plt.show()
model = """
data {
int N;
array[N] real W;
array[N] real E;
}
parameters {
real aW;
real bEW;
real<lower=0> sigma;
}
model {
array[N] real mu;
aW ~ normal(0, 0.2);
bEW ~ normal(0, 0.5);
sigma ~ exponential(1);
for (i in 1:N){
mu[i] = aW + bEW*E[i];
}
W ~ normal(mu, sigma);
}
"""
dat_list = {
'N': len(dat_sim),
'W': dat_sim.W.values,
'E': dat_sim.E.values,
}
posteriori = stan.build(model, data=dat_list)
samples = posteriori.sample(num_chains=4, num_samples=1000)
model_14_4 = az.from_pystan(
posterior=samples,
posterior_model=posteriori,
observed_data=dat_list.keys()
)
az.summary(model_14_4, hdi_prob=0.89)
| mean | sd | hdi_5.5% | hdi_94.5% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| aW | -0.000 | 0.039 | -0.066 | 0.057 | 0.001 | 0.001 | 3968.0 | 2939.0 | 1.0 |
| bEW | 0.413 | 0.040 | 0.349 | 0.479 | 0.001 | 0.000 | 3483.0 | 2369.0 | 1.0 |
| sigma | 0.912 | 0.030 | 0.867 | 0.961 | 0.000 | 0.000 | 4109.0 | 2633.0 | 1.0 |
R Code 14.25¶
model = """
data {
int N;
array[N] real W;
array[N] real E;
array[N] real Q;
}
parameters {
real aW;
real bEW;
real bQW;
real<lower=0> sigma;
}
model {
array[N] real mu;
aW ~ normal(0, 0.2);
bEW ~ normal(0, 0.5);
bQW ~ normal(0, 0.5);
sigma ~ exponential(1);
for (i in 1:N){
mu[i] = aW + bEW*E[i] + bQW*Q[i];
}
W ~ normal(mu, sigma);
}
"""
dat_list = {
'N': len(dat_sim),
'W': dat_sim.W.values,
'E': dat_sim.E.values,
'Q': dat_sim.Q.values,
}
posteriori = stan.build(model, data=dat_list)
samples = posteriori.sample(num_chains=4, num_samples=1000)
model_14_5 = az.from_pystan(
posterior=samples,
posterior_model=posteriori,
observed_data=dat_list.keys()
)
az.summary(model_14_5, hdi_prob=0.89)
| mean | sd | hdi_5.5% | hdi_94.5% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| aW | -0.001 | 0.038 | -0.062 | 0.060 | 0.001 | 0.001 | 3870.0 | 2998.0 | 1.0 |
| bEW | 0.633 | 0.050 | 0.556 | 0.712 | 0.001 | 0.001 | 3293.0 | 2912.0 | 1.0 |
| bQW | -0.348 | 0.050 | -0.421 | -0.257 | 0.001 | 0.001 | 3419.0 | 2644.0 | 1.0 |
| sigma | 0.870 | 0.027 | 0.828 | 0.912 | 0.000 | 0.000 | 3886.0 | 3023.0 | 1.0 |
Include instrument variable \(Q\), the bias increase. The bEW now is \(0.6\) instead of \(0.4\) from previous model. The results from the new model is terrible!
Now, we goes to use instrument variable correctly.
Let’s wirter four models, simples generative versions of the DAG’s.
The firt model for how wages \(W\) are caused by education \(E\) and the unobserverd confound \(U\).
The second model, for education levels \(E\) are caused by quarter of birth \(Q\) and the same unobserved confound \(U\).
The thirt model is for \(Q\), that says that one-quarter of all people born in each quarter of the year.
Finally, the model says that the unobserved confound \(U\) is normally distributed with mean standart gaussian.
Now, we writer this generative model to statistical model, like multivariate normal distribution. Like this: