8 - Os peixes-bois condicionais¶
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import pandas as pd
import networkx as nx
# from causalgraphicalmodels import CausalGraphicalModel
import arviz as az
# ArviZ ships with style sheets!
# https://python.arviz.org/en/stable/examples/styles.html#example-styles
az.style.use("arviz-darkgrid")
import xarray as xr
import stan
import nest_asyncio
plt.style.use('default')
plt.rcParams['axes.facecolor'] = 'lightgray'
# To DAG's
import daft
from causalgraphicalmodels import CausalGraphicalModel
# Add fonts to matplotlib to run xkcd
from matplotlib import font_manager
font_dirs = ["fonts/"] # The path to the custom font file.
font_files = font_manager.findSystemFonts(fontpaths=font_dirs)
for font_file in font_files:
font_manager.fontManager.addfont(font_file)
# plt.xkcd()
# To running the stan in jupyter notebook
nest_asyncio.apply()
R Code 8.1¶
df = pd.read_csv('./data/rugged.csv', sep=';')
df.head()
| isocode | isonum | country | rugged | rugged_popw | rugged_slope | rugged_lsd | rugged_pc | land_area | lat | ... | africa_region_w | africa_region_e | africa_region_c | slave_exports | dist_slavemkt_atlantic | dist_slavemkt_indian | dist_slavemkt_saharan | dist_slavemkt_redsea | pop_1400 | european_descent | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ABW | 533 | Aruba | 0.462 | 0.380 | 1.226 | 0.144 | 0.000 | 18.0 | 12.508 | ... | 0 | 0 | 0 | 0.0 | NaN | NaN | NaN | NaN | 614.0 | NaN |
| 1 | AFG | 4 | Afghanistan | 2.518 | 1.469 | 7.414 | 0.720 | 39.004 | 65209.0 | 33.833 | ... | 0 | 0 | 0 | 0.0 | NaN | NaN | NaN | NaN | 1870829.0 | 0.0 |
| 2 | AGO | 24 | Angola | 0.858 | 0.714 | 2.274 | 0.228 | 4.906 | 124670.0 | -12.299 | ... | 0 | 0 | 1 | 3610000.0 | 5.669 | 6.981 | 4.926 | 3.872 | 1223208.0 | 2.0 |
| 3 | AIA | 660 | Anguilla | 0.013 | 0.010 | 0.026 | 0.006 | 0.000 | 9.0 | 18.231 | ... | 0 | 0 | 0 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | ALB | 8 | Albania | 3.427 | 1.597 | 10.451 | 1.006 | 62.133 | 2740.0 | 41.143 | ... | 0 | 0 | 0 | 0.0 | NaN | NaN | NaN | NaN | 200000.0 | 100.0 |
5 rows × 51 columns
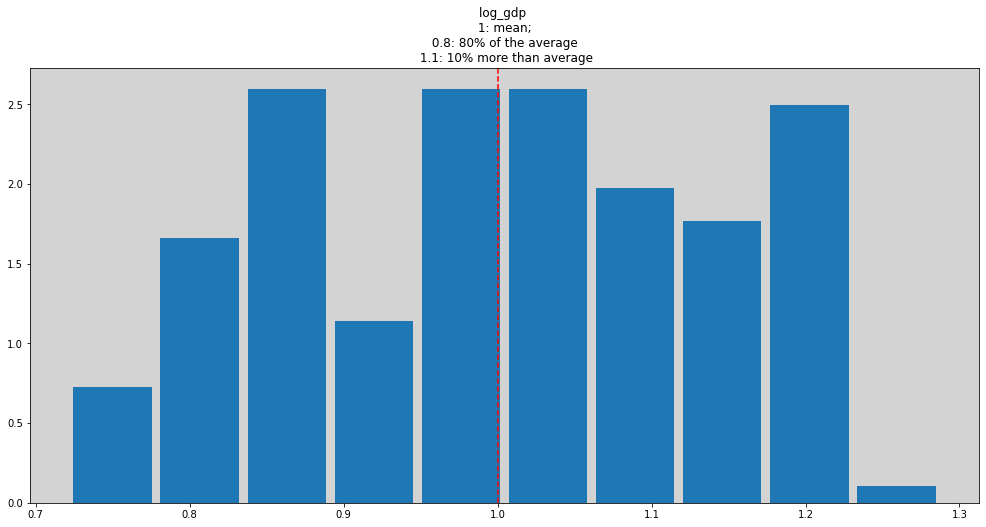
df['log_gdp'] = np.log(df['rgdppc_2000']) # Log version of outcome
df[['rgdppc_2000', 'log_gdp']].head()
| rgdppc_2000 | log_gdp | |
|---|---|---|
| 0 | NaN | NaN |
| 1 | NaN | NaN |
| 2 | 1794.729 | 7.492609 |
| 3 | NaN | NaN |
| 4 | 3703.113 | 8.216929 |
ddf = df[~np.isnan(df['log_gdp'].values)].copy()
ddf['log_gdp_std'] = ddf['log_gdp'] / np.mean(ddf['log_gdp'])
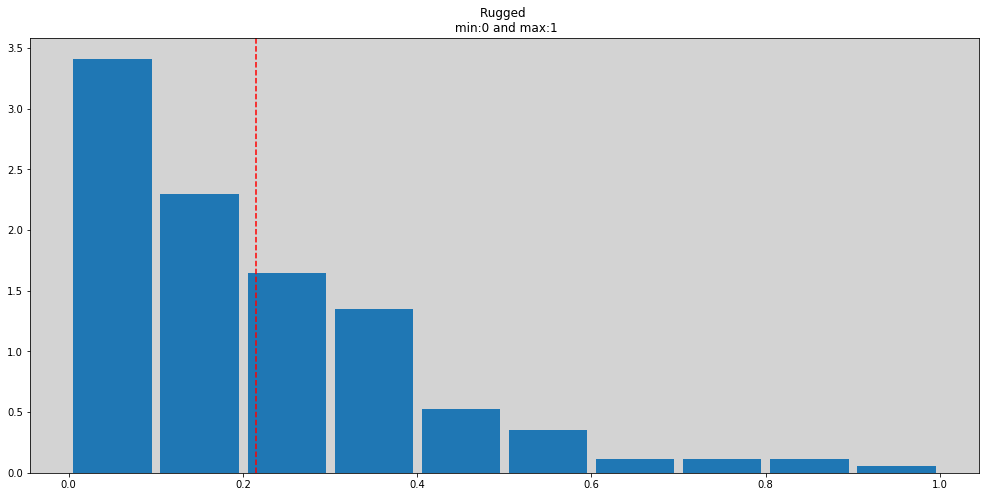
ddf['rugged_std'] = ddf['rugged'] / np.max(ddf['rugged'])
ddf[['log_gdp_std', 'rugged_std']]
| log_gdp_std | rugged_std | |
|---|---|---|
| 2 | 0.879712 | 0.138342 |
| 4 | 0.964755 | 0.552564 |
| 7 | 1.166270 | 0.123992 |
| 8 | 1.104485 | 0.124960 |
| 9 | 0.914904 | 0.433409 |
| ... | ... | ... |
| 229 | 0.996681 | 0.270397 |
| 230 | 0.783032 | 0.374557 |
| 231 | 1.074365 | 0.283941 |
| 232 | 0.780967 | 0.085940 |
| 233 | 0.918589 | 0.192519 |
170 rows × 2 columns
plt.figure(figsize=(17, 8))
plt.hist(ddf['log_gdp_std'], density=True, rwidth=0.9)
plt.title('log_gdp \n 1: mean; \n 0.8: 80% of the average \n 1.1: 10% more than average')
plt.axvline(x=1, c='r', ls='--')
plt.show()
plt.figure(figsize=(17, 8))
plt.hist(ddf['rugged_std'], density=True, rwidth=0.9)
plt.title('Rugged \n min:0 and max:1')
plt.axvline(x=np.mean(ddf['rugged_std']), c='r', ls='--')
plt.show()
R Code 8.2¶
model = """
data {
int N;
vector[N] log_gdp_std;
vector[N] rugged_std;
real rugged_std_average;
}
parameters {
real alpha;
real beta;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta * (rugged_std - rugged_std_average);
}
model {
// Prioris
alpha ~ normal(1, 1);
beta ~ normal(0, 1);
sigma ~ exponential(1);
// Likelihood
log_gdp_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik; // By default, if a variable log_lik is present in the Stan model, it will be retrieved as pointwise log likelihood values.
vector[N] log_gdp_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(log_gdp_std[i] | mu[i], sigma);
log_gdp_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(ddf),
'log_gdp_std': ddf['log_gdp_std'].values,
'rugged_std': ddf['rugged_std'].values,
'rugged_std_average': ddf['rugged_std'].mean(),
}
posteriori = stan.build(model, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
# Transform to dataframe pandas
df_samples = samples.to_frame()
df_samples.head()
| parameters | lp__ | accept_stat__ | stepsize__ | treedepth__ | n_leapfrog__ | divergent__ | energy__ | alpha | beta | sigma | ... | log_gdp_std_hat.161 | log_gdp_std_hat.162 | log_gdp_std_hat.163 | log_gdp_std_hat.164 | log_gdp_std_hat.165 | log_gdp_std_hat.166 | log_gdp_std_hat.167 | log_gdp_std_hat.168 | log_gdp_std_hat.169 | log_gdp_std_hat.170 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| draws | |||||||||||||||||||||
| 0 | 251.095956 | 0.934916 | 0.756421 | 2.0 | 3.0 | 0.0 | -249.133644 | 1.003160 | 0.012665 | 0.132500 | ... | 1.034012 | 0.906565 | 0.989983 | 0.945965 | 1.046566 | 1.012428 | 0.871437 | 0.955593 | 1.041951 | 0.856201 |
| 1 | 247.262579 | 0.989367 | 0.751819 | 2.0 | 3.0 | 0.0 | -246.744846 | 1.005781 | 0.135747 | 0.151454 | ... | 1.044460 | 1.144566 | 0.875533 | 0.860964 | 1.094581 | 1.177379 | 1.083208 | 1.013122 | 0.992572 | 1.167171 |
| 2 | 250.380368 | 0.998516 | 0.943934 | 2.0 | 7.0 | 0.0 | -249.945732 | 1.002515 | 0.032714 | 0.128198 | ... | 1.047150 | 1.007480 | 1.019530 | 1.052204 | 1.112828 | 1.149602 | 1.175402 | 1.279579 | 1.000648 | 0.870344 |
| 3 | 250.516786 | 0.913792 | 0.862509 | 2.0 | 3.0 | 0.0 | -249.646704 | 1.002620 | 0.045010 | 0.130002 | ... | 1.144794 | 0.994287 | 1.006064 | 1.133213 | 0.878218 | 1.246487 | 0.956800 | 0.669329 | 1.084075 | 0.973772 |
| 4 | 247.927975 | 0.574694 | 0.756421 | 2.0 | 3.0 | 0.0 | -246.256142 | 1.018086 | -0.077998 | 0.129151 | ... | 0.921305 | 0.814742 | 0.918643 | 0.982626 | 0.949444 | 0.886278 | 1.159055 | 0.902815 | 0.837451 | 1.115775 |
5 rows × 520 columns
# stan_fit to arviz_stan
stan_data = az.from_pystan(
posterior=samples,
posterior_predictive="log_gdp_std_hat",
observed_data=['log_gdp_std'],
prior=samples,
prior_model=posteriori,
posterior_model=posteriori,
)
stan_data
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 7 ... 163 164 165 166 167 168 169 Data variables: alpha (chain, draw) float64 1.003 1.018 1.01 ... 1.0 0.9874 0.9897 beta (chain, draw) float64 0.01267 -0.078 -0.09028 ... -0.06006 -0.0592 sigma (chain, draw) float64 0.1325 0.1292 0.1447 ... 0.1439 0.1427 mu (chain, draw, mu_dim_0) float64 1.002 1.007 1.002 ... 0.9973 0.991 Attributes: created_at: 2023-08-11T19:31:12.686511 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/tsiivt35 program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 1.119... Attributes: created_at: 2023-08-11T19:31:12.851711 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_lik_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 ... 164 165 166 167 168 169 Data variables: log_lik (chain, draw, log_lik_dim_0) float64 0.675 1.05 ... 0.899 Attributes: created_at: 2023-08-11T19:31:12.800781 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9349 0.5747 0.946 ... 0.9162 1.0 step_size (chain, draw) float64 0.7564 0.7564 ... 0.8625 0.8625 tree_depth (chain, draw) int64 2 2 2 2 2 2 2 3 2 ... 2 3 2 3 3 2 2 2 1 n_steps (chain, draw) int64 3 3 3 3 3 3 3 7 3 ... 3 7 3 7 7 3 3 7 1 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -249.1 -246.3 ... -247.2 -249.7 lp (chain, draw) float64 251.1 247.9 249.2 ... 249.7 250.1 Attributes: created_at: 2023-08-11T19:31:12.739805 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/tsiivt35 program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 170, log_lik_dim_0: 170, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 ... 165 166 167 168 169 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: alpha (chain, draw) float64 1.003 1.018 ... 0.9874 0.9897 beta (chain, draw) float64 0.01267 -0.078 ... -0.0592 sigma (chain, draw) float64 0.1325 0.1292 ... 0.1439 0.1427 mu (chain, draw, mu_dim_0) float64 1.002 1.007 ... 0.991 log_lik (chain, draw, log_lik_dim_0) float64 0.675 ... 0.899 log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 1.119... Attributes: created_at: 2023-08-11T19:31:12.909230 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/tsiivt35 program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: lp (chain, draw) float64 251.1 247.9 249.2 ... 249.7 250.1 acceptance_rate (chain, draw) float64 0.9349 0.5747 0.946 ... 0.9162 1.0 step_size (chain, draw) float64 0.7564 0.7564 ... 0.8625 0.8625 tree_depth (chain, draw) int64 2 2 2 2 2 2 2 3 2 ... 2 3 2 3 3 2 2 2 1 n_steps (chain, draw) int64 3 3 3 3 3 3 3 7 3 ... 3 7 3 7 7 3 3 7 1 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -249.1 -246.3 ... -247.2 -249.7 Attributes: created_at: 2023-08-11T19:31:12.966971 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/tsiivt35 program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (log_gdp_std_dim_0: 170) Coordinates: * log_gdp_std_dim_0 (log_gdp_std_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 Data variables: log_gdp_std (log_gdp_std_dim_0) float64 0.8797 0.9648 ... 0.9186 Attributes: created_at: 2023-08-11T19:31:12.653606 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0
az.summary(stan_data, var_names=['alpha', 'beta', 'sigma'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 1.000 | 0.010 | 0.980 | 1.019 | 0.000 | 0.000 | 4227.0 | 2654.0 | 1.0 |
| beta | 0.002 | 0.056 | -0.097 | 0.113 | 0.001 | 0.001 | 3467.0 | 2810.0 | 1.0 |
| sigma | 0.138 | 0.008 | 0.125 | 0.153 | 0.000 | 0.000 | 3838.0 | 2808.0 | 1.0 |
R Code 8.3¶
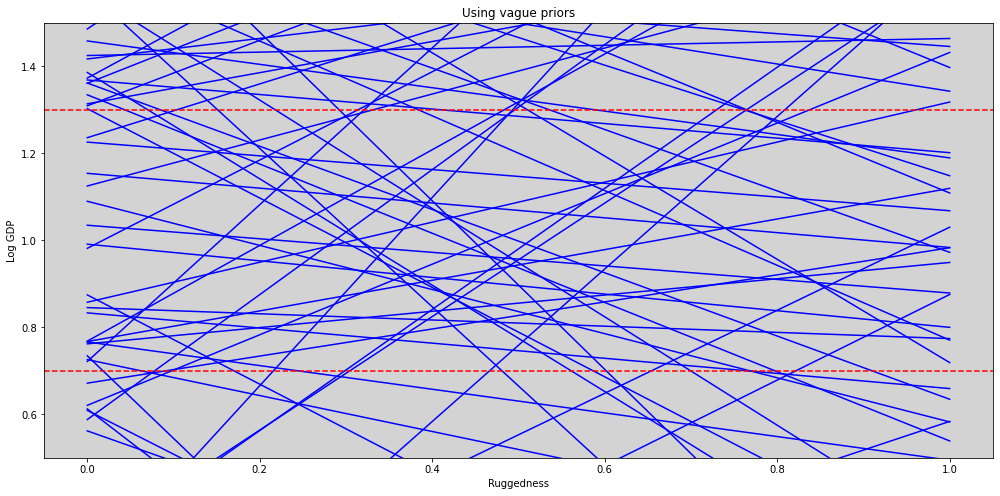
plt.figure(figsize=(17, 8))
alpha = np.random.normal(1, 1, 1000)
beta = np.random.normal(0, 1, 1000)
rugged_seq = np.linspace(0, 1, 100)
for i in range(100):
plt.plot(rugged_seq, alpha[i] + beta[i] * rugged_seq, c='blue')
plt.axhline(y=1.3, c='r', ls='--')
plt.axhline(y=0.7, c='r', ls='--')
plt.ylim((0.5, 1.5))
plt.title('Using vague priors')
plt.xlabel('Ruggedness')
plt.ylabel('Log GDP')
plt.show()
R Code 8.4¶
plt.figure(figsize=(17, 8))
plt.hist(beta, bins=30, rwidth=0.9)
plt.axvline(x=0.6, c='r', ls='--')
plt.axvline(x=-0.6, c='r', ls='--')
plt.show()
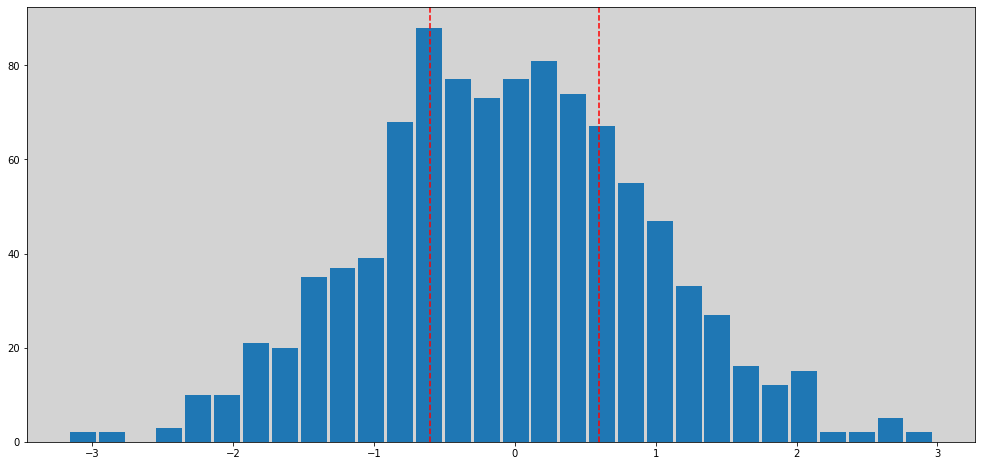
np.sum(np.sum(np.abs(beta) > 0.6) / len(beta))
0.55
R Code 8.5¶
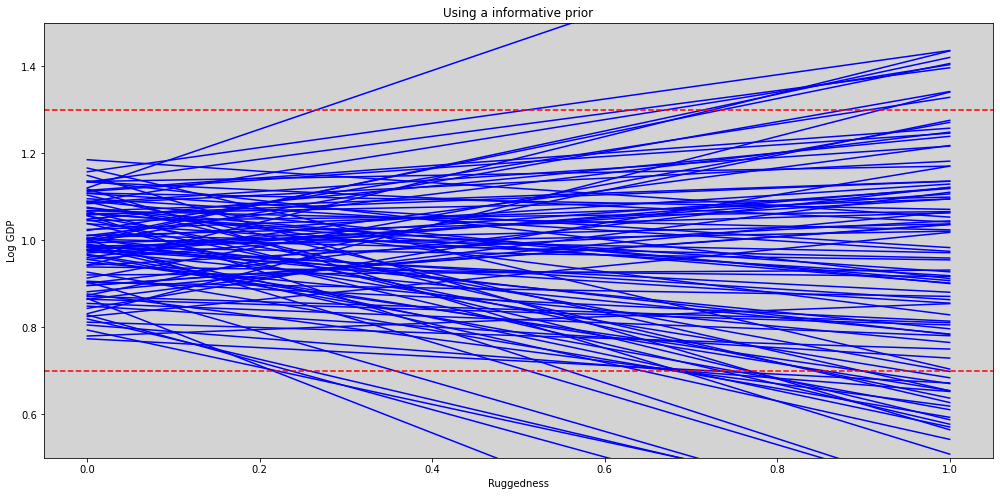
plt.figure(figsize=(17, 8))
alpha = np.random.normal(1, 0.1, 1000)
beta = np.random.normal(0, 0.3, 1000)
rugged_seq = np.linspace(0, 1, 100)
for i in range(100):
plt.plot(rugged_seq, alpha[i] + beta[i] * rugged_seq, c='blue')
plt.axhline(y=1.3, c='r', ls='--')
plt.axhline(y=0.7, c='r', ls='--')
plt.ylim((0.5, 1.5))
plt.title('Using a informative prior')
plt.xlabel('Ruggedness')
plt.ylabel('Log GDP')
plt.show()
model2 = """
data {
int N;
vector[N] log_gdp_std;
vector[N] rugged_std;
real rugged_std_average;
}
parameters {
real alpha;
real beta;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta * (rugged_std - rugged_std_average);
}
model {
// Prioris
alpha ~ normal(1, 0.1);
beta ~ normal(0, 0.3);
sigma ~ exponential(1);
// Likelihood
log_gdp_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik; // By default, if a variable log_lik is present in the Stan model, it will be retrieved as pointwise log likelihood values.
vector[N] log_gdp_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(log_gdp_std[i] | mu[i], sigma);
log_gdp_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(ddf),
'log_gdp_std': ddf['log_gdp_std'].values,
'rugged_std': ddf['rugged_std'].values,
'rugged_std_average': ddf['rugged_std'].mean(),
}
posteriori = stan.build(model2, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
stan_data2 = az.from_pystan(
posterior=samples,
posterior_predictive="log_gdp_std_hat",
observed_data=['log_gdp_std'],
prior=samples,
prior_model=posteriori,
posterior_model=posteriori,
)
az.summary(stan_data, var_names=['alpha', 'beta', 'sigma'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 1.000 | 0.010 | 0.980 | 1.019 | 0.000 | 0.000 | 4227.0 | 2654.0 | 1.0 |
| beta | 0.002 | 0.056 | -0.097 | 0.113 | 0.001 | 0.001 | 3467.0 | 2810.0 | 1.0 |
| sigma | 0.138 | 0.008 | 0.125 | 0.153 | 0.000 | 0.000 | 3838.0 | 2808.0 | 1.0 |
R Code 8.7¶
ddf['cid'] = [1 if cont_africa == 1 else 2 for cont_africa in ddf['cont_africa']]
ddf[['cont_africa', 'cid']].head()
| cont_africa | cid | |
|---|---|---|
| 2 | 1 | 1 |
| 4 | 0 | 2 |
| 7 | 0 | 2 |
| 8 | 0 | 2 |
| 9 | 0 | 2 |
R Code 8.8¶
model3 = """
data {
int N;
vector[N] log_gdp_std;
vector[N] rugged_std;
array[N] int cid; // Must be integer because this is index to alpha.
real rugged_std_average;
}
parameters {
real alpha[2]; //Can be used to real alpha[2] or array[2] int alpha;
real beta;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
for (i in 1:N){
mu[i] = alpha[ cid[i] ] + beta * (rugged_std[i] - rugged_std_average);
}
}
model {
// Prioris
alpha ~ normal(1, 0.1);
beta ~ normal(0, 0.3);
sigma ~ exponential(1);
// Likelihood
log_gdp_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] log_gdp_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(log_gdp_std[i] | mu[i], sigma);
log_gdp_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(ddf),
'log_gdp_std': ddf['log_gdp_std'].values,
'rugged_std': ddf['rugged_std'].values,
'rugged_std_average': ddf['rugged_std'].mean(),
'cid': ddf['cid'].values,
}
posteriori = stan.build(model3, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
stan_data3 = az.from_pystan(
posterior=samples,
posterior_predictive="log_gdp_std_hat",
observed_data=['log_gdp_std'],
prior=samples,
prior_model=posteriori,
posterior_model=posteriori,
dims={
"alpha": ["africa"],
},
)
R Code 8.9¶
models_8 = { 'm8.1': stan_data2, 'm8.2': stan_data3 }
az.compare(models_8, ic='waic')
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/stats.py:1645: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(
| rank | elpd_waic | p_waic | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| m8.2 | 0 | 126.166124 | 4.117261 | 0.000000 | 0.969256 | 7.396644 | 0.000000 | True | log |
| m8.1 | 1 | 94.499399 | 2.491680 | 31.666725 | 0.030744 | 6.463956 | 7.317875 | False | log |
R Code 8.10¶
az.summary(stan_data3, var_names=['alpha', 'beta', 'sigma'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha[0] | 0.880 | 0.016 | 0.851 | 0.911 | 0.000 | 0.000 | 4512.0 | 3483.0 | 1.0 |
| alpha[1] | 1.049 | 0.010 | 1.031 | 1.069 | 0.000 | 0.000 | 4305.0 | 2642.0 | 1.0 |
| beta | -0.046 | 0.047 | -0.130 | 0.048 | 0.001 | 0.001 | 3508.0 | 2517.0 | 1.0 |
| sigma | 0.114 | 0.006 | 0.103 | 0.126 | 0.000 | 0.000 | 3742.0 | 3076.0 | 1.0 |
R Code 8.11¶
stan_data3
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, africa: 2, mu_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * africa (africa) int64 0 1 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 7 ... 163 164 165 166 167 168 169 Data variables: alpha (chain, draw, africa) float64 0.8731 1.052 0.8843 ... 0.8777 1.072 beta (chain, draw) float64 -0.04755 -0.02791 ... -0.07995 0.03227 sigma (chain, draw) float64 0.1152 0.1109 0.11 ... 0.1056 0.1162 0.1162 mu (chain, draw, mu_dim_0) float64 0.8768 1.036 ... 0.8736 0.877 Attributes: created_at: 2023-08-11T18:56:27.122181 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/22jq2cue program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 0.746... Attributes: created_at: 2023-08-11T18:56:27.276036 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_lik_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 ... 164 165 166 167 168 169 Data variables: log_lik (chain, draw, log_lik_dim_0) float64 1.242 1.051 ... 1.169 Attributes: created_at: 2023-08-11T18:56:27.225035 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9829 0.9857 0.9661 ... 0.7619 1.0 step_size (chain, draw) float64 0.7249 0.7249 ... 0.7518 0.7518 tree_depth (chain, draw) int64 3 3 2 2 3 2 3 2 3 ... 3 2 3 2 3 3 3 3 3 n_steps (chain, draw) int64 7 7 3 7 7 3 7 3 7 ... 7 3 7 3 7 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -281.1 -282.8 ... -274.9 -275.5 lp (chain, draw) float64 283.2 283.2 282.8 ... 278.0 279.2 Attributes: created_at: 2023-08-11T18:56:27.172412 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/22jq2cue program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, africa: 2, mu_dim_0: 170, log_lik_dim_0: 170, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * africa (africa) int64 0 1 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 ... 165 166 167 168 169 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: alpha (chain, draw, africa) float64 0.8731 1.052 ... 1.072 beta (chain, draw) float64 -0.04755 -0.02791 ... 0.03227 sigma (chain, draw) float64 0.1152 0.1109 ... 0.1162 0.1162 mu (chain, draw, mu_dim_0) float64 0.8768 ... 0.877 log_lik (chain, draw, log_lik_dim_0) float64 1.242 ... 1.169 log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 0.746... Attributes: created_at: 2023-08-11T18:56:27.342979 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/22jq2cue program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: lp (chain, draw) float64 283.2 283.2 282.8 ... 278.0 279.2 acceptance_rate (chain, draw) float64 0.9829 0.9857 0.9661 ... 0.7619 1.0 step_size (chain, draw) float64 0.7249 0.7249 ... 0.7518 0.7518 tree_depth (chain, draw) int64 3 3 2 2 3 2 3 2 3 ... 3 2 3 2 3 3 3 3 3 n_steps (chain, draw) int64 7 7 3 7 7 3 7 3 7 ... 7 3 7 3 7 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -281.1 -282.8 ... -274.9 -275.5 Attributes: created_at: 2023-08-11T18:56:27.392039 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/22jq2cue program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (log_gdp_std_dim_0: 170) Coordinates: * log_gdp_std_dim_0 (log_gdp_std_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 Data variables: log_gdp_std (log_gdp_std_dim_0) float64 0.8797 0.9648 ... 0.9186 Attributes: created_at: 2023-08-11T18:56:27.082048 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0
alpha_a1 = stan_data3.posterior.alpha.sel(africa=0)
alpha_a2 = stan_data3.posterior.alpha.sel(africa=1)
diff_alpha_a1_a2 = az.extract(alpha_a1 - alpha_a2).alpha.values
az.hdi(diff_alpha_a1_a2, hdi_prob=0.89)
array([-0.19925646, -0.13838364])
R Code 8.12¶
# Extract 200 samples from arviz-fit to numpy
params_post = az.extract(stan_data3.posterior, num_samples=200)
rugged_seq = np.linspace(0, 1, 30)
log_gdp_mean_africa = []
log_gdp_hdi_africa = []
log_gdp_mean_not_africa = []
log_gdp_hdi_not_africa = []
# Calculation posterior mean and interval HDI
for i in range(len(rugged_seq)):
log_gdp_africa = params_post.alpha.sel(africa=0) + params_post.beta.values * rugged_seq[i]
log_gdp_mean_africa.append(np.mean(log_gdp_africa.values))
log_gdp_hdi_africa.append(az.hdi(log_gdp_africa.values, hdi_prob=0.89))
log_gdp_not_africa = params_post.alpha.sel(africa=1) + params_post.beta.values * rugged_seq[i]
log_gdp_mean_not_africa.append(np.mean(log_gdp_not_africa.values))
log_gdp_hdi_not_africa.append(az.hdi(log_gdp_not_africa.values, hdi_prob=0.89))
log_gdp_hdi_africa = np.array(log_gdp_hdi_africa)
log_gdp_hdi_not_africa = np.array(log_gdp_hdi_not_africa)
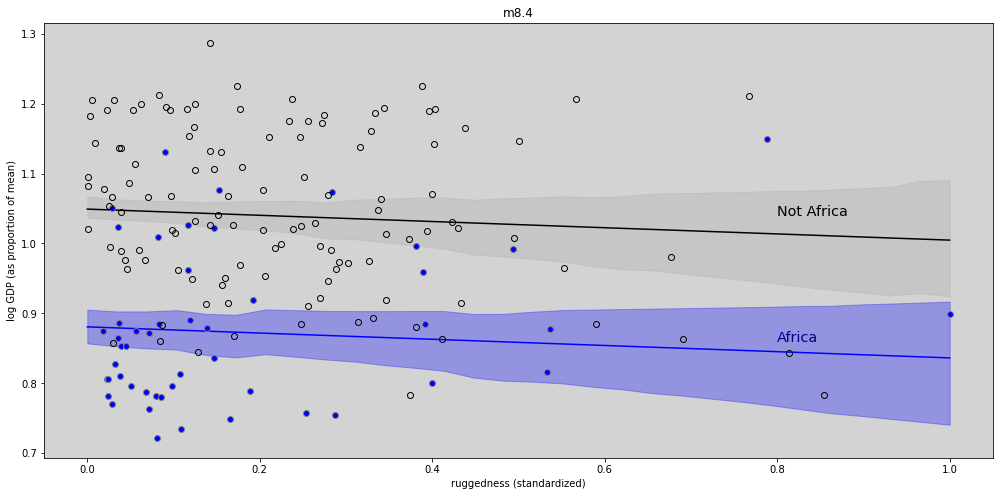
plt.figure(figsize=(17, 8))
plt.plot(rugged_seq, log_gdp_mean_africa, c='blue')
plt.fill_between(rugged_seq, log_gdp_hdi_africa[:,0], log_gdp_hdi_africa[:,1], color='blue', alpha=0.3)
plt.plot(rugged_seq, log_gdp_mean_not_africa, c='black')
plt.fill_between(rugged_seq, log_gdp_hdi_not_africa[:,0], log_gdp_hdi_not_africa[:,1], color='darkgray', alpha=0.3)
plt.plot(ddf.loc[ddf.cid == 1,'rugged_std'], ddf.loc[ddf.cid==1, 'log_gdp_std'], 'o', markerfacecolor='blue', color='gray')
plt.plot(ddf.loc[ddf.cid == 2,'rugged_std'], ddf.loc[ddf.cid==2, 'log_gdp_std'], 'o', markerfacecolor='none', color='black')
plt.title('m8.4')
plt.xlabel('ruggedness (standardized)')
plt.ylabel('log GDP (as proportion of mean)')
plt.text(0.8, 1.04, 'Not Africa', fontsize='x-large', color='black')
plt.text(0.8, 0.86, 'Africa', fontsize='x-large', color='darkblue')
plt.show()
R Code 8.13¶
model4 = """
data {
int N;
vector[N] log_gdp_std;
vector[N] rugged_std;
array[N] int cid; // Must be integer because this is index to alpha.
real rugged_std_average;
}
parameters {
real alpha[2]; //Can be used to real alpha[2] or array[2] int alpha;
real beta[2];
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
for (i in 1:N){
mu[i] = alpha[ cid[i] ] + beta[ cid[i] ] * (rugged_std[i] - rugged_std_average);
}
}
model {
// Prioris
alpha ~ normal(1, 0.1);
beta ~ normal(0, 0.3);
sigma ~ exponential(1);
// Likelihood
log_gdp_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] log_gdp_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(log_gdp_std[i] | mu[i], sigma);
log_gdp_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(ddf),
'log_gdp_std': ddf['log_gdp_std'].values,
'rugged_std': ddf['rugged_std'].values,
'rugged_std_average': ddf['rugged_std'].mean(),
'cid': ddf['cid'].values,
}
posteriori = stan.build(model4, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
stan_data4 = az.from_pystan(
posterior=samples,
posterior_predictive="log_gdp_std_hat",
observed_data=['log_gdp_std'],
prior=samples,
prior_model=posteriori,
posterior_model=posteriori,
dims={
"alpha": ["africa"],
"beta": ["africa"],
},
)
R Code 8.14¶
az.summary(stan_data4, var_names=['alpha', 'beta', 'sigma'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha[0] | 0.887 | 0.016 | 0.858 | 0.916 | 0.000 | 0.000 | 5156.0 | 2914.0 | 1.0 |
| alpha[1] | 1.051 | 0.010 | 1.033 | 1.071 | 0.000 | 0.000 | 5063.0 | 3095.0 | 1.0 |
| beta[0] | 0.131 | 0.075 | -0.006 | 0.270 | 0.001 | 0.001 | 4710.0 | 3386.0 | 1.0 |
| beta[1] | -0.143 | 0.056 | -0.247 | -0.036 | 0.001 | 0.001 | 4727.0 | 3117.0 | 1.0 |
| sigma | 0.112 | 0.006 | 0.100 | 0.123 | 0.000 | 0.000 | 5069.0 | 3262.0 | 1.0 |
R Code 8.15¶
models_8 = { 'm8.1': stan_data2, 'm8.2': stan_data3, 'm8.3': stan_data4 }
# https://python.arviz.org/en/stable/api/generated/arviz.compare.html
# https://rss.onlinelibrary.wiley.com/doi/10.1111/1467-9868.00353
az.compare(models_8, ic='loo') # loo is same that PSIS
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| m8.3 | 0 | 129.589277 | 4.989499 | 0.000000 | 0.870793 | 7.309693 | 0.000000 | False | log |
| m8.2 | 1 | 126.137495 | 4.145890 | 3.451783 | 0.129207 | 7.405339 | 3.225278 | False | log |
| m8.1 | 2 | 94.492463 | 2.498616 | 35.096814 | 0.000000 | 6.464447 | 7.448111 | False | log |
R Code 8.16¶
az.loo(stan_data4, pointwise=True)
Computed from 4000 posterior samples and 170 observations log-likelihood matrix.
Estimate SE
elpd_loo 129.59 7.31
p_loo 4.99 -
------
Pareto k diagnostic values:
Count Pct.
(-Inf, 0.5] (good) 169 99.4%
(0.5, 0.7] (ok) 1 0.6%
(0.7, 1] (bad) 0 0.0%
(1, Inf) (very bad) 0 0.0%
stan_data4
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, africa: 2, mu_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * africa (africa) int64 0 1 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 7 ... 163 164 165 166 167 168 169 Data variables: alpha (chain, draw, africa) float64 0.88 1.051 0.8771 ... 0.876 1.022 beta (chain, draw, africa) float64 0.2193 -0.1387 ... 0.1464 -0.1622 sigma (chain, draw) float64 0.1212 0.131 0.1102 ... 0.1118 0.1209 0.1094 mu (chain, draw, mu_dim_0) float64 0.8632 1.004 ... 0.8572 0.8728 Attributes: created_at: 2023-08-11T18:56:30.413832 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/5jmion4o program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 0.913... Attributes: created_at: 2023-08-11T18:56:30.571354 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_lik_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 ... 164 165 166 167 168 169 Data variables: log_lik (chain, draw, log_lik_dim_0) float64 1.182 1.139 ... 1.206 Attributes: created_at: 2023-08-11T18:56:30.520437 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9562 0.7774 ... 0.9232 0.8468 step_size (chain, draw) float64 0.7482 0.7482 ... 0.6294 0.6294 tree_depth (chain, draw) int64 2 3 3 3 2 3 3 3 2 ... 3 3 2 3 3 3 3 2 3 n_steps (chain, draw) int64 3 7 7 7 3 7 7 7 3 ... 7 7 7 7 7 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -280.2 -279.4 ... -284.5 -279.4 lp (chain, draw) float64 285.4 280.7 282.5 ... 285.7 283.1 Attributes: created_at: 2023-08-11T18:56:30.463167 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/5jmion4o program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, africa: 2, mu_dim_0: 170, log_lik_dim_0: 170, log_gdp_std_hat_dim_0: 170) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 ... 994 995 996 997 998 999 * africa (africa) int64 0 1 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 ... 165 166 167 168 169 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 * log_gdp_std_hat_dim_0 (log_gdp_std_hat_dim_0) int64 0 1 2 3 ... 167 168 169 Data variables: alpha (chain, draw, africa) float64 0.88 1.051 ... 1.022 beta (chain, draw, africa) float64 0.2193 ... -0.1622 sigma (chain, draw) float64 0.1212 0.131 ... 0.1209 0.1094 mu (chain, draw, mu_dim_0) float64 0.8632 ... 0.8728 log_lik (chain, draw, log_lik_dim_0) float64 1.182 ... 1.206 log_gdp_std_hat (chain, draw, log_gdp_std_hat_dim_0) float64 0.913... Attributes: created_at: 2023-08-11T18:56:30.647322 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/5jmion4o program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: lp (chain, draw) float64 285.4 280.7 282.5 ... 285.7 283.1 acceptance_rate (chain, draw) float64 0.9562 0.7774 ... 0.9232 0.8468 step_size (chain, draw) float64 0.7482 0.7482 ... 0.6294 0.6294 tree_depth (chain, draw) int64 2 3 3 3 2 3 3 3 2 ... 3 3 2 3 3 3 3 2 3 n_steps (chain, draw) int64 3 7 7 7 3 7 7 7 3 ... 7 7 7 7 7 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -280.2 -279.4 ... -284.5 -279.4 Attributes: created_at: 2023-08-11T18:56:30.696630 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/5jmion4o program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (log_gdp_std_dim_0: 170) Coordinates: * log_gdp_std_dim_0 (log_gdp_std_dim_0) int64 0 1 2 3 4 ... 166 167 168 169 Data variables: log_gdp_std (log_gdp_std_dim_0) float64 0.8797 0.9648 ... 0.9186 Attributes: created_at: 2023-08-11T18:56:30.365835 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0
R Code 8.16 - XXX¶
# az.plot_loo_pit(stan_data4, y='log_gdp_std')
R Code 8.17¶
# Extract 200 samples from arviz-fit to numpy
params_post = az.extract(stan_data4.posterior, num_samples=200)
rugged_seq = np.linspace(0, 1, 30)
log_gdp_mean_africa = []
log_gdp_hdi_africa = []
log_gdp_mean_not_africa = []
log_gdp_hdi_not_africa = []
# Calculation posterior mean and interval HDI
for i in range(len(rugged_seq)):
log_gdp_africa = params_post.alpha.sel(africa=0) + params_post.beta.sel(africa=0).values * rugged_seq[i]
log_gdp_mean_africa.append(np.mean(log_gdp_africa.values))
log_gdp_hdi_africa.append(az.hdi(log_gdp_africa.values, hdi_prob=0.89))
log_gdp_not_africa = params_post.alpha.sel(africa=1) + params_post.beta.sel(africa=1).values * rugged_seq[i]
log_gdp_mean_not_africa.append(np.mean(log_gdp_not_africa.values))
log_gdp_hdi_not_africa.append(az.hdi(log_gdp_not_africa.values, hdi_prob=0.89))
log_gdp_hdi_africa = np.array(log_gdp_hdi_africa)
log_gdp_hdi_not_africa = np.array(log_gdp_hdi_not_africa)
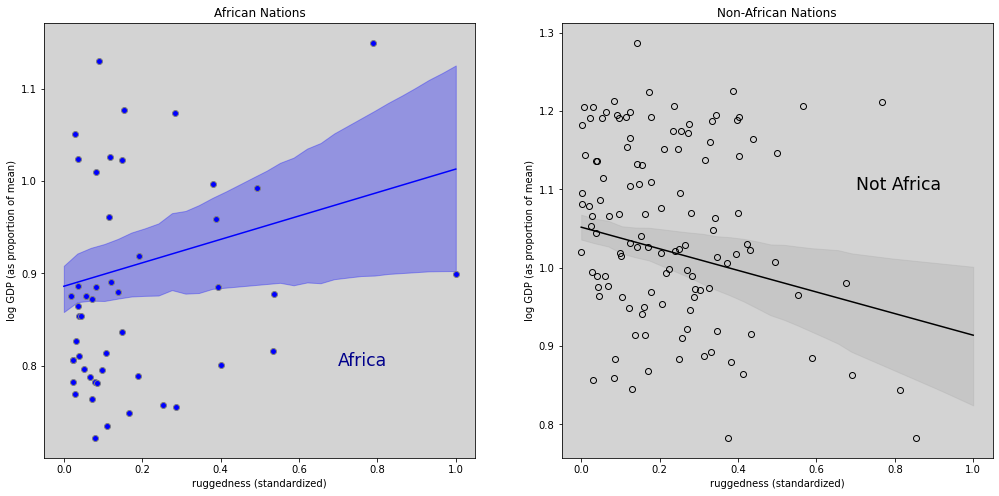
fig = plt.figure(figsize=(17, 8))
gs = GridSpec(1, 2)
# Africa plots
ax_africa = fig.add_subplot(gs[0])
ax_africa.plot(rugged_seq, log_gdp_mean_africa, c='blue')
ax_africa.fill_between(rugged_seq, log_gdp_hdi_africa[:,0], log_gdp_hdi_africa[:,1], color='blue', alpha=0.3)
ax_africa.plot(ddf.loc[ddf.cid == 1,'rugged_std'], ddf.loc[ddf.cid==1, 'log_gdp_std'], 'o', markerfacecolor='blue', color='gray')
ax_africa.set_title('African Nations')
ax_africa.set_xlabel('ruggedness (standardized)')
ax_africa.set_ylabel('log GDP (as proportion of mean)')
ax_africa.text(0.7, 0.8, 'Africa', fontsize='xx-large', color='darkblue')
# Non africa plots
ax_not_africa = fig.add_subplot(gs[1])
ax_not_africa.plot(rugged_seq, log_gdp_mean_not_africa, c='black')
ax_not_africa.fill_between(rugged_seq, log_gdp_hdi_not_africa[:,0], log_gdp_hdi_not_africa[:,1], color='darkgray', alpha=0.3)
ax_not_africa.plot(ddf.loc[ddf.cid == 2,'rugged_std'], ddf.loc[ddf.cid==2, 'log_gdp_std'], 'o', markerfacecolor='none', color='black')
ax_not_africa.set_title('Non-African Nations')
ax_not_africa.set_xlabel('ruggedness (standardized)')
ax_not_africa.set_ylabel('log GDP (as proportion of mean)')
ax_not_africa.text(0.7, 1.1, 'Not Africa', fontsize='xx-large', color='black')
plt.show()
R Code 8.18¶
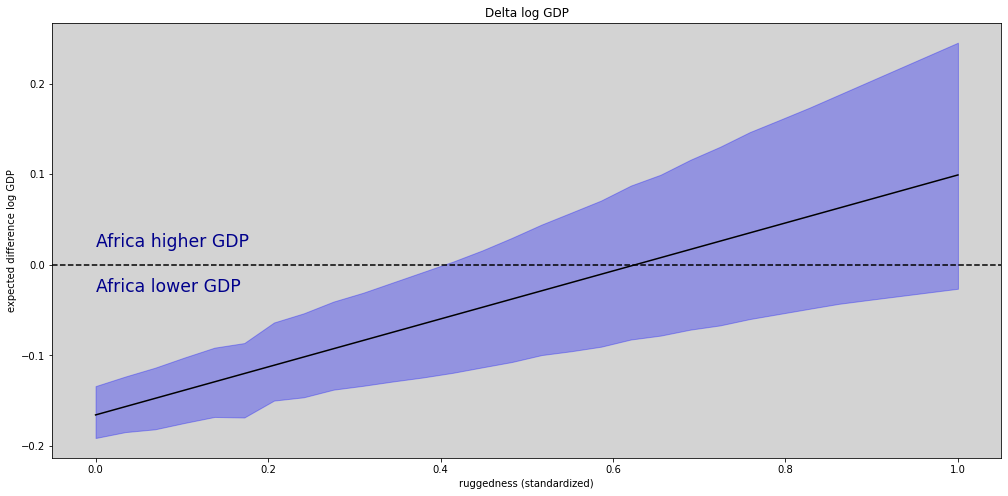
log_gdp_delta_mean = []
log_gdp_delta_hdi = []
# Calculation posterior mean and interval HDI
for i in range(len(rugged_seq)):
log_gdp_africa = params_post.alpha.sel(africa=0) + params_post.beta.sel(africa=0).values * rugged_seq[i]
log_gdp_not_africa = params_post.alpha.sel(africa=1) + params_post.beta.sel(africa=1).values * rugged_seq[i]
log_gdp_delta = log_gdp_africa - log_gdp_not_africa
log_gdp_delta_mean.append(np.mean(log_gdp_delta.values))
log_gdp_delta_hdi.append(az.hdi(log_gdp_delta.values, hdi_prob=0.89))
log_gdp_delta_hdi = np.array(log_gdp_delta_hdi)
fig = plt.figure(figsize=(17, 8))
gs = GridSpec(1, 1)
# Africa delta log GDP plots
ax_delta = fig.add_subplot(gs[0])
ax_delta.plot(rugged_seq, log_gdp_delta_mean, c='black')
ax_delta.fill_between(rugged_seq, log_gdp_delta_hdi[:,0], log_gdp_delta_hdi[:,1], color='blue', alpha=0.3)
ax_delta.set_title('Delta log GDP')
ax_delta.set_xlabel('ruggedness (standardized)')
ax_delta.set_ylabel('expected difference log GDP')
ax_delta.text(0.0, 0.02, 'Africa higher GDP', fontsize='xx-large', color='darkblue')
ax_delta.text(0.0, -0.03, 'Africa lower GDP', fontsize='xx-large', color='darkblue')
ax_delta.axhline(y=0, ls='--', color='black')
plt.show()
R Code 8.19¶
df = pd.read_csv('./data/tulips.csv', sep=';',
dtype={
'bed': 'category', # cluster of plants from same section of the greenhouse
'water': 'float', # Predictor: Soil moisture - (1) low and (3) high
'shade': 'float', # Predictor: Light exposure - (1) high and (3) low
'blooms': 'float', # What we wish to predict
})
df.tail()
| bed | water | shade | blooms | |
|---|---|---|---|---|
| 22 | c | 2.0 | 2.0 | 135.92 |
| 23 | c | 2.0 | 3.0 | 90.66 |
| 24 | c | 3.0 | 1.0 | 304.52 |
| 25 | c | 3.0 | 2.0 | 249.33 |
| 26 | c | 3.0 | 3.0 | 134.59 |
df.describe(include='category')
| bed | |
|---|---|
| count | 27 |
| unique | 3 |
| top | a |
| freq | 9 |
df.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| water | 27.0 | 2.000000 | 0.832050 | 1.0 | 1.000 | 2.00 | 3.0 | 3.00 |
| shade | 27.0 | 2.000000 | 0.832050 | 1.0 | 1.000 | 2.00 | 3.0 | 3.00 |
| blooms | 27.0 | 128.993704 | 92.683923 | 0.0 | 71.115 | 111.04 | 190.3 | 361.66 |
R Code 8.20¶
df['blooms_std'] = df['blooms'] / df['blooms'].max()
df['water_cent'] = df['water'] - df['water'].mean()
df['shade_cent'] = df['shade'] - df['shade'].mean()
R Code 8.21¶
\[ \alpha \sim Normal(0.5, 1) \]
alpha = np.random.normal(0.5, 1, 1000)
alphas_true = np.any([alpha < 0, alpha > 1], axis=0) # If (alpha < 0) or (alpha > 1) then True else False
np.sum(alphas_true) / len(alpha)
0.628
R Code 8.22¶
\[ \alpha \sim Normal(0.5, 0.25) \]
alpha = np.random.normal(0.5, 0.25, 1000)
alphas_true = np.any([alpha < 0, alpha > 1], axis=0) # If (alpha < 0) or (alpha > 1) then True else False
np.sum(alphas_true) / len(alpha)
0.053
R Code 8.23¶
model = """
data {
int N;
vector[N] blooms_std;
vector[N] water_cent;
vector[N] shade_cent;
}
parameters {
real alpha;
real beta_w;
real beta_s;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta_w * water_cent + beta_s * shade_cent;
}
model {
// Priori
alpha ~ normal(0.5, 0.25);
beta_w ~ normal(0, 0.25);
beta_s ~ normal(0, 0.25);
sigma ~ exponential(1);
blooms_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] blooms_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(blooms_std[i] | mu[i], sigma);
blooms_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(df),
'blooms_std': df.blooms_std.values,
'water_cent': df.water_cent.values,
'shade_cent': df.shade_cent.values,
}
posteriori = stan.build(model, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
stan_blooms = az.from_pystan(
prior_model=posteriori,
prior=samples,
posterior_model=posteriori,
posterior=samples,
posterior_predictive="blooms_std_hat",
observed_data=['blooms_std', 'water_cent', 'shade_cent'],
)
stan_blooms
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 7 8 ... 18 19 20 21 22 23 24 25 26 Data variables: alpha (chain, draw) float64 0.3615 0.3282 0.3282 ... 0.3661 0.3922 0.34 beta_w (chain, draw) float64 0.2235 0.2231 0.2231 ... 0.2044 0.2055 beta_s (chain, draw) float64 -0.08126 -0.07431 ... -0.1841 -0.04058 sigma (chain, draw) float64 0.1818 0.1567 0.1567 ... 0.1719 0.1567 mu (chain, draw, mu_dim_0) float64 0.2193 0.1381 ... 0.5455 0.505 Attributes: created_at: 2023-08-11T18:56:33.510826 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/elkgjo5c program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, blooms_std_hat_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 994 995 996 997 998 999 * blooms_std_hat_dim_0 (blooms_std_hat_dim_0) int64 0 1 2 3 4 ... 23 24 25 26 Data variables: blooms_std_hat (chain, draw, blooms_std_hat_dim_0) float64 0.1343 ... Attributes: created_at: 2023-08-11T18:56:33.654050 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_lik_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 6 ... 20 21 22 23 24 25 26 Data variables: log_lik (chain, draw, log_lik_dim_0) float64 0.05831 ... 0.5753 Attributes: created_at: 2023-08-11T18:56:33.609401 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9282 0.9287 ... 0.8313 0.9974 step_size (chain, draw) float64 0.804 0.804 0.804 ... 0.6157 0.6157 tree_depth (chain, draw) int64 2 2 2 3 2 1 3 3 2 ... 2 2 3 3 2 2 3 3 3 n_steps (chain, draw) int64 3 3 3 7 3 3 7 7 3 ... 7 7 7 7 3 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -32.69 -31.76 -25.78 ... -28.81 -30.5 lp (chain, draw) float64 32.9 32.41 32.41 ... 31.39 31.47 Attributes: created_at: 2023-08-11T18:56:33.561609 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/elkgjo5c program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 27, log_lik_dim_0: 27, blooms_std_hat_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 ... 21 22 23 24 25 26 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 ... 22 23 24 25 26 * blooms_std_hat_dim_0 (blooms_std_hat_dim_0) int64 0 1 2 3 4 ... 23 24 25 26 Data variables: alpha (chain, draw) float64 0.3615 0.3282 ... 0.3922 0.34 beta_w (chain, draw) float64 0.2235 0.2231 ... 0.2044 0.2055 beta_s (chain, draw) float64 -0.08126 -0.07431 ... -0.04058 sigma (chain, draw) float64 0.1818 0.1567 ... 0.1719 0.1567 mu (chain, draw, mu_dim_0) float64 0.2193 ... 0.505 log_lik (chain, draw, log_lik_dim_0) float64 0.05831 ... 0.... blooms_std_hat (chain, draw, blooms_std_hat_dim_0) float64 0.1343 ... Attributes: created_at: 2023-08-11T18:56:33.701908 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/elkgjo5c program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: lp (chain, draw) float64 32.9 32.41 32.41 ... 31.39 31.47 acceptance_rate (chain, draw) float64 0.9282 0.9287 ... 0.8313 0.9974 step_size (chain, draw) float64 0.804 0.804 0.804 ... 0.6157 0.6157 tree_depth (chain, draw) int64 2 2 2 3 2 1 3 3 2 ... 2 2 3 3 2 2 3 3 3 n_steps (chain, draw) int64 3 3 3 7 3 3 7 7 3 ... 7 7 7 7 3 7 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -32.69 -31.76 -25.78 ... -28.81 -30.5 Attributes: created_at: 2023-08-11T18:56:33.750637 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/elkgjo5c program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (blooms_std_dim_0: 27, water_cent_dim_0: 27, shade_cent_dim_0: 27) Coordinates: * blooms_std_dim_0 (blooms_std_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 * water_cent_dim_0 (water_cent_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 * shade_cent_dim_0 (shade_cent_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 Data variables: blooms_std (blooms_std_dim_0) float64 0.0 0.0 0.307 ... 0.6894 0.3721 water_cent (water_cent_dim_0) float64 -1.0 -1.0 -1.0 ... 1.0 1.0 1.0 shade_cent (shade_cent_dim_0) float64 -1.0 0.0 1.0 ... -1.0 0.0 1.0 Attributes: created_at: 2023-08-11T18:56:33.478728 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0
az.summary(stan_blooms, var_names=['alpha', 'beta_w', 'beta_s'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.359 | 0.034 | 0.298 | 0.429 | 0.001 | 0.0 | 3857.0 | 2579.0 | 1.0 |
| beta_w | 0.203 | 0.041 | 0.126 | 0.279 | 0.001 | 0.0 | 4353.0 | 3032.0 | 1.0 |
| beta_s | -0.112 | 0.041 | -0.188 | -0.036 | 0.001 | 0.0 | 4151.0 | 2524.0 | 1.0 |
R Code 8.24¶
\[ B_i \sim Normal(\mu_i, \sigma) \]
\[ \mu_i = \alpha + \beta_W W_i + \beta_S S_i + \beta_{WS} S_i W_i \]
model = """
data {
int N;
vector[N] blooms_std;
vector[N] water_cent;
vector[N] shade_cent;
}
parameters {
real alpha;
real beta_w;
real beta_s;
real beta_ws;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta_w * water_cent + beta_s * shade_cent + beta_ws * water_cent .* shade_cent;
}
model {
// Priori
alpha ~ normal(0.5, 0.25);
beta_w ~ normal(0, 0.25);
beta_s ~ normal(0, 0.25);
beta_ws ~ normal(0, 0.25);
sigma ~ exponential(1);
blooms_std ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] blooms_std_hat;
for(i in 1:N){
log_lik[i] = normal_lpdf(blooms_std[i] | mu[i], sigma);
blooms_std_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(df),
'blooms_std': df.blooms_std.values,
'water_cent': df.water_cent.values,
'shade_cent': df.shade_cent.values,
}
posteriori = stan.build(model, data=data)
samples = posteriori.sample(num_chains=4, num_samples=1000)
stan_blooms_interaction = az.from_pystan(
prior_model=posteriori,
prior=samples,
posterior_model=posteriori,
posterior=samples,
posterior_predictive="blooms_std_hat",
observed_data=['blooms_std', 'water_cent', 'shade_cent'],
)
stan_blooms_interaction
arviz.InferenceData
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 8 ... 992 993 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 7 8 ... 18 19 20 21 22 23 24 25 26 Data variables: alpha (chain, draw) float64 0.3674 0.3462 0.354 ... 0.3529 0.3582 0.3258 beta_w (chain, draw) float64 0.2106 0.2189 0.1748 ... 0.2551 0.2051 beta_s (chain, draw) float64 -0.1585 -0.1735 ... -0.1014 -0.06658 beta_ws (chain, draw) float64 -0.2024 -0.1429 -0.1649 ... -0.08582 -0.1958 sigma (chain, draw) float64 0.1466 0.1574 0.1383 ... 0.1805 0.1231 mu (chain, draw, mu_dim_0) float64 0.1129 0.1568 ... 0.5309 0.2686 Attributes: created_at: 2023-08-11T18:56:34.758903 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/2akx274x program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, blooms_std_hat_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 994 995 996 997 998 999 * blooms_std_hat_dim_0 (blooms_std_hat_dim_0) int64 0 1 2 3 4 ... 23 24 25 26 Data variables: blooms_std_hat (chain, draw, blooms_std_hat_dim_0) float64 0.04633... Attributes: created_at: 2023-08-11T18:56:34.902871 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, log_lik_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 6 ... 20 21 22 23 24 25 26 Data variables: log_lik (chain, draw, log_lik_dim_0) float64 0.7045 0.4295 ... 0.8218 Attributes: created_at: 2023-08-11T18:56:34.858158 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9824 0.9818 ... 0.9986 0.9993 step_size (chain, draw) float64 0.7264 0.7264 ... 0.6313 0.6313 tree_depth (chain, draw) int64 2 2 3 3 2 2 3 3 2 ... 3 3 2 3 3 2 3 3 3 n_steps (chain, draw) int64 7 3 7 7 3 3 7 7 3 ... 7 7 3 7 7 3 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -36.43 -35.68 ... -32.65 -33.83 lp (chain, draw) float64 37.17 37.08 37.86 ... 35.73 36.25 Attributes: created_at: 2023-08-11T18:56:34.810343 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/2akx274x program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, mu_dim_0: 27, log_lik_dim_0: 27, blooms_std_hat_dim_0: 27) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 994 995 996 997 998 999 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 ... 21 22 23 24 25 26 * log_lik_dim_0 (log_lik_dim_0) int64 0 1 2 3 4 5 ... 22 23 24 25 26 * blooms_std_hat_dim_0 (blooms_std_hat_dim_0) int64 0 1 2 3 4 ... 23 24 25 26 Data variables: alpha (chain, draw) float64 0.3674 0.3462 ... 0.3582 0.3258 beta_w (chain, draw) float64 0.2106 0.2189 ... 0.2551 0.2051 beta_s (chain, draw) float64 -0.1585 -0.1735 ... -0.06658 beta_ws (chain, draw) float64 -0.2024 -0.1429 ... -0.1958 sigma (chain, draw) float64 0.1466 0.1574 ... 0.1805 0.1231 mu (chain, draw, mu_dim_0) float64 0.1129 ... 0.2686 log_lik (chain, draw, log_lik_dim_0) float64 0.7045 ... 0.8218 blooms_std_hat (chain, draw, blooms_std_hat_dim_0) float64 0.04633... Attributes: created_at: 2023-08-11T18:56:34.950864 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/2akx274x program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: lp (chain, draw) float64 37.17 37.08 37.86 ... 35.73 36.25 acceptance_rate (chain, draw) float64 0.9824 0.9818 ... 0.9986 0.9993 step_size (chain, draw) float64 0.7264 0.7264 ... 0.6313 0.6313 tree_depth (chain, draw) int64 2 2 3 3 2 2 3 3 2 ... 3 3 2 3 3 2 3 3 3 n_steps (chain, draw) int64 7 3 7 7 3 3 7 7 3 ... 7 7 3 7 7 3 7 7 7 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 -36.43 -35.68 ... -32.65 -33.83 Attributes: created_at: 2023-08-11T18:56:35.000044 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 model_name: models/2akx274x program_code: \n data {\n int N;\n vector[... random_seed: None -
<xarray.Dataset> Dimensions: (blooms_std_dim_0: 27, water_cent_dim_0: 27, shade_cent_dim_0: 27) Coordinates: * blooms_std_dim_0 (blooms_std_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 * water_cent_dim_0 (water_cent_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 * shade_cent_dim_0 (shade_cent_dim_0) int64 0 1 2 3 4 5 ... 21 22 23 24 25 26 Data variables: blooms_std (blooms_std_dim_0) float64 0.0 0.0 0.307 ... 0.6894 0.3721 water_cent (water_cent_dim_0) float64 -1.0 -1.0 -1.0 ... 1.0 1.0 1.0 shade_cent (shade_cent_dim_0) float64 -1.0 0.0 1.0 ... -1.0 0.0 1.0 Attributes: created_at: 2023-08-11T18:56:34.723805 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0
az.summary(stan_blooms_interaction, var_names=['alpha', 'beta_w', 'beta_s', 'beta_ws'])
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| alpha | 0.358 | 0.029 | 0.301 | 0.411 | 0.000 | 0.0 | 5629.0 | 2730.0 | 1.0 |
| beta_w | 0.206 | 0.034 | 0.145 | 0.275 | 0.000 | 0.0 | 4954.0 | 2852.0 | 1.0 |
| beta_s | -0.113 | 0.034 | -0.177 | -0.051 | 0.000 | 0.0 | 5101.0 | 3033.0 | 1.0 |
| beta_ws | -0.143 | 0.042 | -0.220 | -0.059 | 0.001 | 0.0 | 5011.0 | 3012.0 | 1.0 |
R Code 8.25¶
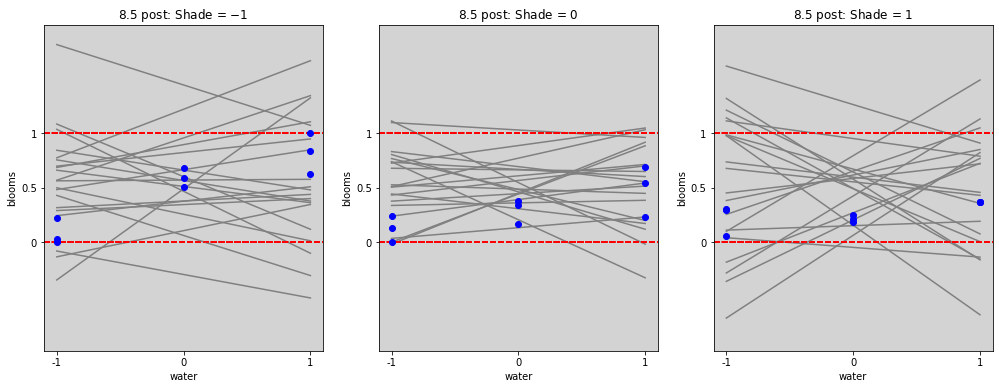
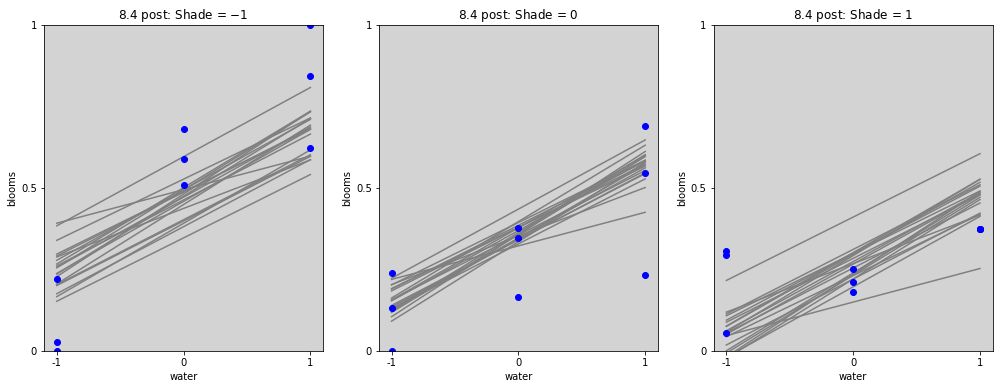
blooms_post = az.extract(stan_blooms.posterior, num_samples=20) # get 20 lines
blooms_post_int = az.extract(stan_blooms_interaction.posterior, num_samples=20) # get 20 lines
# ================
# Plot without interactions
water_seq = np.linspace(-1, 1, 30)
fig = plt.figure(figsize=(17, 6))
gs = GridSpec(1, 3)
shade_cent_values = [-1, 0, 1]
ax = [None] * len(shade_cent_values)
for j, shade_plot_aux in enumerate(shade_cent_values):
ax[j] = fig.add_subplot(gs[j])
for i in range(len(blooms_post.alpha)):
mu = blooms_post.alpha[i].values + \
blooms_post.beta_w[i].values * water_seq + \
blooms_post.beta_s[i].values * (shade_plot_aux)
ax[j].plot(water_seq, mu, c='gray')
ax[j].set_ylim(0, 1)
ax[j].set_title(f'8.4 post: Shade = ${shade_plot_aux}$')
ax[j].set_xlabel('water')
ax[j].set_ylabel('blooms')
ax[j].set_xticks(shade_cent_values, shade_cent_values)
ax[j].set_yticks([0, 0.5, 1.0], [0, 0.5, 1])
ax[j].plot(
df.loc[df['shade_cent'] == shade_plot_aux,'water_cent'].values,
df.loc[df['shade_cent'] == shade_plot_aux,'blooms_std'].values,
'o', c='blue')
plt.show()
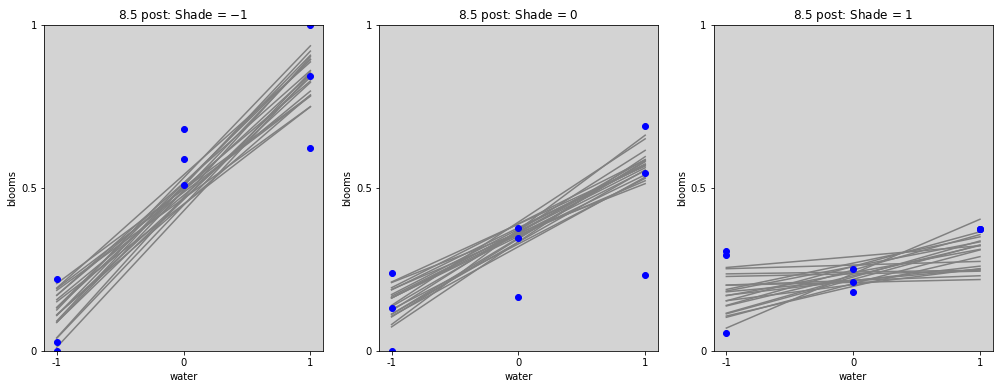
# ================
# Plot with interactions
water_seq = np.linspace(-1, 1, 30)
fig = plt.figure(figsize=(17, 6))
gs = GridSpec(1, 3)
shade_cent_values = [-1, 0, 1]
ax = [None] * len(shade_cent_values)
for j, shade_plot_aux in enumerate(shade_cent_values):
ax[j] = fig.add_subplot(gs[j])
for i in range(len(blooms_post_int.alpha)):
mu = blooms_post_int.alpha[i].values + \
blooms_post_int.beta_w[i].values * water_seq + \
blooms_post_int.beta_s[i].values * (shade_plot_aux) + \
blooms_post_int.beta_ws[i].values * water_seq * (shade_plot_aux)
ax[j].plot(water_seq, mu, c='gray')
ax[j].set_ylim(0, 1)
ax[j].set_title(f'8.5 post: Shade = ${shade_plot_aux}$')
ax[j].set_xlabel('water')
ax[j].set_ylabel('blooms')
ax[j].set_xticks(shade_cent_values, shade_cent_values)
ax[j].set_yticks([0, 0.5, 1.0], [0, 0.5, 1])
ax[j].plot(
df.loc[df['shade_cent'] == shade_plot_aux,'water_cent'].values,
df.loc[df['shade_cent'] == shade_plot_aux,'blooms_std'].values,
'o', c='blue')
plt.show()
R Code 8.26¶
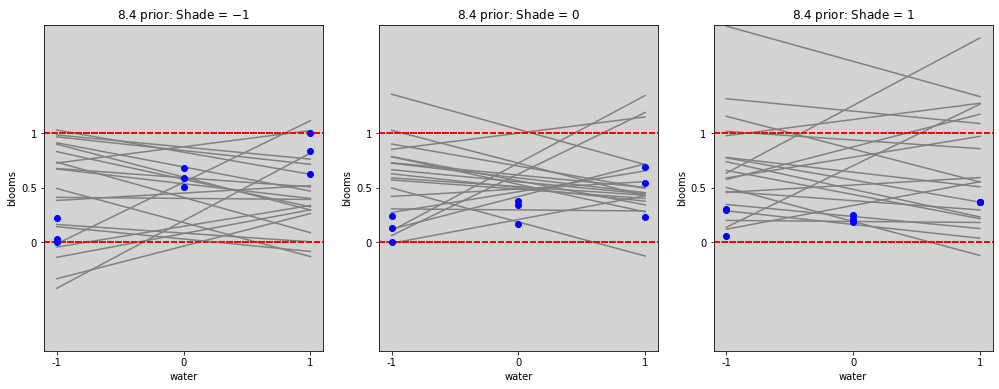
# ================
# Plot without interactions
# Prior
alpha_prior = np.random.normal(0.5, 0.25, 20)
beta_w_prior = np.random.normal(0, 0.25, 20)
beta_s_prior = np.random.normal(0, 0.25, 20)
water_seq = np.linspace(-1, 1, 30)
fig = plt.figure(figsize=(17, 6))
gs = GridSpec(1, 3)
shade_cent_values = [-1, 0, 1]
ax = [None] * len(shade_cent_values)
for j, shade_plot_aux in enumerate(shade_cent_values):
ax[j] = fig.add_subplot(gs[j])
for i in range(len(alpha_prior)):
mu = alpha_prior[i] + \
beta_w_prior[i] * water_seq + \
beta_s_prior[i] * (shade_plot_aux)
ax[j].plot(water_seq, mu, c='gray')
ax[j].set_ylim(0, 1)
ax[j].set_title(f'8.4 prior: Shade = ${shade_plot_aux}$')
ax[j].set_xlabel('water')
ax[j].set_ylabel('blooms')
ax[j].set_xticks(shade_cent_values, shade_cent_values)
ax[j].set_yticks([0, 0.5, 1.0], [0, 0.5, 1])
ax[j].set_ylim(-1, 2)
ax[j].axhline(y=1, ls='--', color='red')
ax[j].axhline(y=0, ls='--', color='red')
ax[j].plot(
df.loc[df['shade_cent'] == shade_plot_aux,'water_cent'].values,
df.loc[df['shade_cent'] == shade_plot_aux,'blooms_std'].values,
'o', c='blue')
plt.show()
# ================
# Plot with interactions
# Prior
alpha_prior = np.random.normal(0.5, 0.25, 20)
beta_w_prior = np.random.normal(0, 0.25, 20)
beta_s_prior = np.random.normal(0, 0.25, 20)
beta_ws_prior = np.random.normal(0, 0.25, 20)
water_seq = np.linspace(-1, 1, 30)
fig = plt.figure(figsize=(17, 6))
gs = GridSpec(1, 3)
shade_cent_values = [-1, 0, 1]
ax = [None] * len(shade_cent_values)
for j, shade_plot_aux in enumerate(shade_cent_values):
ax[j] = fig.add_subplot(gs[j])
for i in range(len(alpha_prior)):
mu = alpha_prior[i] + \
beta_w_prior[i] * water_seq + \
beta_s_prior[i] * (shade_plot_aux) + \
beta_ws_prior[i] * water_seq * (shade_plot_aux)
ax[j].plot(water_seq, mu, c='gray')
ax[j].set_ylim(0, 1)
ax[j].set_title(f'8.5 post: Shade = ${shade_plot_aux}$')
ax[j].set_xlabel('water')
ax[j].set_ylabel('blooms')
ax[j].set_xticks(shade_cent_values, shade_cent_values)
ax[j].set_yticks([0, 0.5, 1.0], [0, 0.5, 1])
ax[j].set_ylim(-1, 2)
ax[j].axhline(y=1, ls='--', color='red')
ax[j].axhline(y=0, ls='--', color='red')
ax[j].plot(
df.loc[df['shade_cent'] == shade_plot_aux,'water_cent'].values,
df.loc[df['shade_cent'] == shade_plot_aux,'blooms_std'].values,
'o', c='blue')
plt.show()