7 - Compasso de Ulisses¶
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import pandas as pd
from videpy import Vide
import networkx as nx
# from causalgraphicalmodels import CausalGraphicalModel
import stan
import nest_asyncio
plt.style.use('default')
plt.rcParams['axes.facecolor'] = 'lightgray'
# To DAG's
import daft
from causalgraphicalmodels import CausalGraphicalModel
# To running the stan in jupyter notebook
nest_asyncio.apply()
R Code 7.1 - Pag 194¶
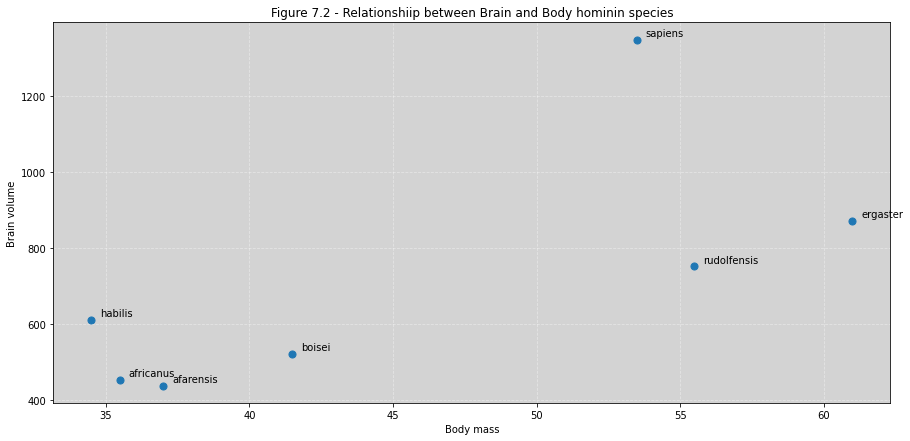
sppnames = ('afarensis', 'africanus', 'habilis', 'boisei', 'rudolfensis', 'ergaster', 'sapiens')
brainvolcc = (438, 452, 612, 521, 752, 871, 1350)
masskg = (37.0, 35.5, 34.5, 41.5, 55.5, 61.0, 53.5)
d = pd.DataFrame({'species':sppnames, 'brain': brainvolcc, 'mass':masskg})
d
| species | brain | mass | |
|---|---|---|---|
| 0 | afarensis | 438 | 37.0 |
| 1 | africanus | 452 | 35.5 |
| 2 | habilis | 612 | 34.5 |
| 3 | boisei | 521 | 41.5 |
| 4 | rudolfensis | 752 | 55.5 |
| 5 | ergaster | 871 | 61.0 |
| 6 | sapiens | 1350 | 53.5 |
fig, ax = plt.subplots(figsize=(15, 7))
ax.scatter(d.mass, d.brain, marker='o', s=50)
for i, text in enumerate(sppnames):
ax.annotate(text, (d.mass[i]+0.3, d.brain[i]+10))
ax.set_title('Figure 7.2 - Relationshiip between Brain and Body hominin species')
ax.set_xlabel('Body mass')
ax.set_ylabel('Brain volume')
ax.grid(ls='--', color='white', alpha=0.4)
plt.show()
R Code 7.2 - pag 196¶
np.std(d.mass, ddof=1)
10.90489186863706
d.mass.std()
10.90489186863706
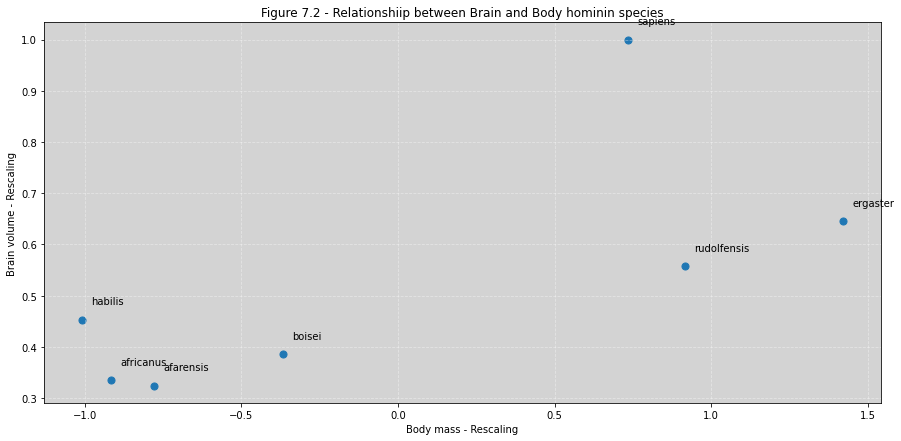
d['mass_std'] = (d.mass - d.mass.mean())/d.mass.std()
d['brain_std'] = d.brain / np.max(d.brain)
d
| species | brain | mass | mass_std | brain_std | |
|---|---|---|---|---|---|
| 0 | afarensis | 438 | 37.0 | -0.779467 | 0.324444 |
| 1 | africanus | 452 | 35.5 | -0.917020 | 0.334815 |
| 2 | habilis | 612 | 34.5 | -1.008722 | 0.453333 |
| 3 | boisei | 521 | 41.5 | -0.366808 | 0.385926 |
| 4 | rudolfensis | 752 | 55.5 | 0.917020 | 0.557037 |
| 5 | ergaster | 871 | 61.0 | 1.421380 | 0.645185 |
| 6 | sapiens | 1350 | 53.5 | 0.733616 | 1.000000 |
fig, ax = plt.subplots(figsize=(15, 7))
ax.scatter(d.mass_std, d.brain_std, marker='o', s=50)
for i, text in enumerate(sppnames):
ax.annotate(text, (d.mass_std[i]+0.03, d.brain_std[i]+0.03))
ax.set_title('Figure 7.2 - Relationshiip between Brain and Body hominin species')
ax.set_xlabel('Body mass - Rescaling')
ax.set_ylabel('Brain volume - Rescaling')
ax.grid(ls='--', color='white', alpha=0.4)
plt.show()
R Code 7.3 - pag 196¶
Modelo linear:
\(\begin{align} b_i \sim Normal(\mu_i, \sigma) \\ \mu_i = \alpha + \beta m_i \\ \alpha \sim Normal(0.5, 1) \\ \beta \sim Normal(0, 10) \\ \sigma \sim LogNormal(0, 1) \\ \end{align}\)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta;
real<lower=0> sigma;
// real log_sigma; // Like the book
}
model {
vector[N] mu;
mu = alpha + beta * body;
// Prioris
alpha ~ normal(0.5, 1);
beta ~ normal(0, 10);
sigma ~ lognormal(0, 1);
// log_sigma ~ normal(0, 1); // Like the book
// Likelihood
brain ~ normal(mu, sigma);
// brain ~ normal(mu, exp(sigma)); // Like the book
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_1 = stan.build(model, data=data)
samples_1 = posteriori_1.sample(num_chains=4, num_samples=1000)
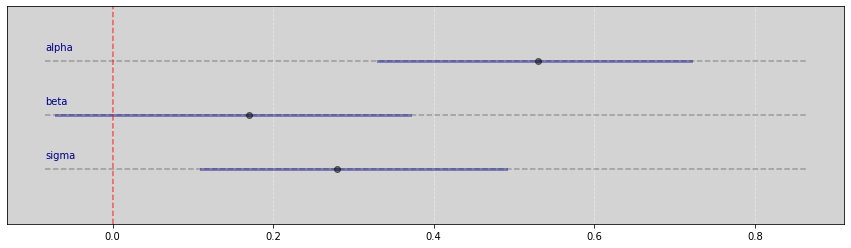
Vide.summary(samples_1)
| mean | std | 7.0% | 93.0% | |
|---|---|---|---|---|
| alpha | 0.53 | 0.11 | 0.33 | 0.72 |
| beta | 0.17 | 0.12 | -0.07 | 0.37 |
| sigma | 0.28 | 0.12 | 0.11 | 0.49 |
Vide.plot_forest(samples_1)
R Code 7.5 - pag 197¶
def var2(x):
return np.sum(np.power(x - np.mean(x), 2))/len(x)
mean_std = [np.mean((samples_1['alpha'].flatten() + samples_1['beta'].flatten() * mass)) for mass in d.mass_std.values]
r = mean_std - d.brain_std
resid_var = var2(r)
outcome_var = var2(d.brain_std)
1 - resid_var/outcome_var
0.490158020661996
R Code 7.6 - Pag 197¶
def R2_is_bad():
pass
R Code 7.7 - pag 198¶
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[2];
real<lower=0> sigma;
}
model {
vector[N] mu;
mu = alpha + beta[1] * body + beta[2] * body^2;
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:2){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 1);
// Likelihood
brain ~ normal(mu, sigma);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_2 = stan.build(model, data=data)
samples_2 = posteriori_2.sample(num_chains=4, num_samples=1000)
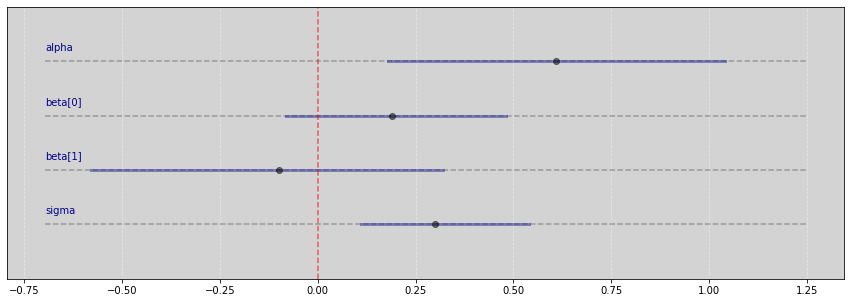
Vide.summary(samples_2)
| mean | std | 7.0% | 93.0% | |
|---|---|---|---|---|
| alpha | 0.61 | 0.24 | 0.18 | 1.04 |
| beta[0] | 0.19 | 0.16 | -0.08 | 0.48 |
| beta[1] | -0.10 | 0.24 | -0.58 | 0.32 |
| sigma | 0.30 | 0.16 | 0.11 | 0.54 |
Vide.plot_forest(samples_2)
R Code 7.8 - Pag 198¶
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[3];
real<lower=0> sigma;
}
model {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3;
// Likelihood
brain ~ normal(mu, sigma);
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:3){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 1);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_3 = stan.build(model, data=data)
samples_3 = posteriori_3.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[4];
real<lower=0> sigma;
}
model {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4;
// Likelihood
brain ~ normal(mu, sigma);
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:4){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 1);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_4 = stan.build(model, data=data)
samples_4 = posteriori_4.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[5];
real<lower=0> sigma;
}
model {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4 +
beta[5] * body^5;
// Likelihood
brain ~ normal(mu, sigma);
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:5){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 1);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_5 = stan.build(model, data=data)
samples_5 = posteriori_5.sample(num_chains=4, num_samples=1000)
R Code 7.9 - Pag 199¶
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[6];
}
model {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4 +
beta[5] * body^5 +
beta[6] * body^6;
// Likelihood
brain ~ normal(mu, 0.001);
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:6){
beta[j] ~ normal(0, 10);
}
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_6 = stan.build(model, data=data)
samples_6 = posteriori_6.sample(num_chains=4, num_samples=1000)
R Code 7.10 - Pag 199¶
mass_seq = np.linspace(start=d.mass_std.min(), stop=d.mass_std.max(), num=100)
pp_1 = [samples_1['alpha'].flatten() + samples_1['beta'].flatten() * mass_i for mass_i in mass_seq]
pp_2 = [samples_2['alpha'].flatten() +
samples_2['beta'][0].flatten() * mass_i +
samples_2['beta'][1].flatten() * np.power(mass_i, 2) for mass_i in mass_seq]
pp_3 = [samples_3['alpha'].flatten() +
samples_3['beta'][0].flatten() * mass_i +
samples_3['beta'][1].flatten() * np.power(mass_i, 2) +
samples_3['beta'][2].flatten() * np.power(mass_i, 3)
for mass_i in mass_seq]
pp_4 = [samples_4['alpha'].flatten() +
samples_4['beta'][0].flatten() * mass_i +
samples_4['beta'][1].flatten() * np.power(mass_i, 2) +
samples_4['beta'][2].flatten() * np.power(mass_i, 3) +
samples_4['beta'][3].flatten() * np.power(mass_i, 4)
for mass_i in mass_seq]
pp_5 = [samples_5['alpha'].flatten() +
samples_5['beta'][0].flatten() * mass_i +
samples_5['beta'][1].flatten() * np.power(mass_i, 2) +
samples_5['beta'][2].flatten() * np.power(mass_i, 3) +
samples_5['beta'][3].flatten() * np.power(mass_i, 4) +
samples_5['beta'][4].flatten() * np.power(mass_i, 5)
for mass_i in mass_seq]
pp_6 = [samples_6['alpha'].flatten() +
samples_6['beta'][0].flatten() * mass_i +
samples_6['beta'][1].flatten() * np.power(mass_i, 2) +
samples_6['beta'][2].flatten() * np.power(mass_i, 3) +
samples_6['beta'][3].flatten() * np.power(mass_i, 4) +
samples_6['beta'][4].flatten() * np.power(mass_i, 5) +
samples_6['beta'][5].flatten() * np.power(mass_i, 6)
for mass_i in mass_seq]
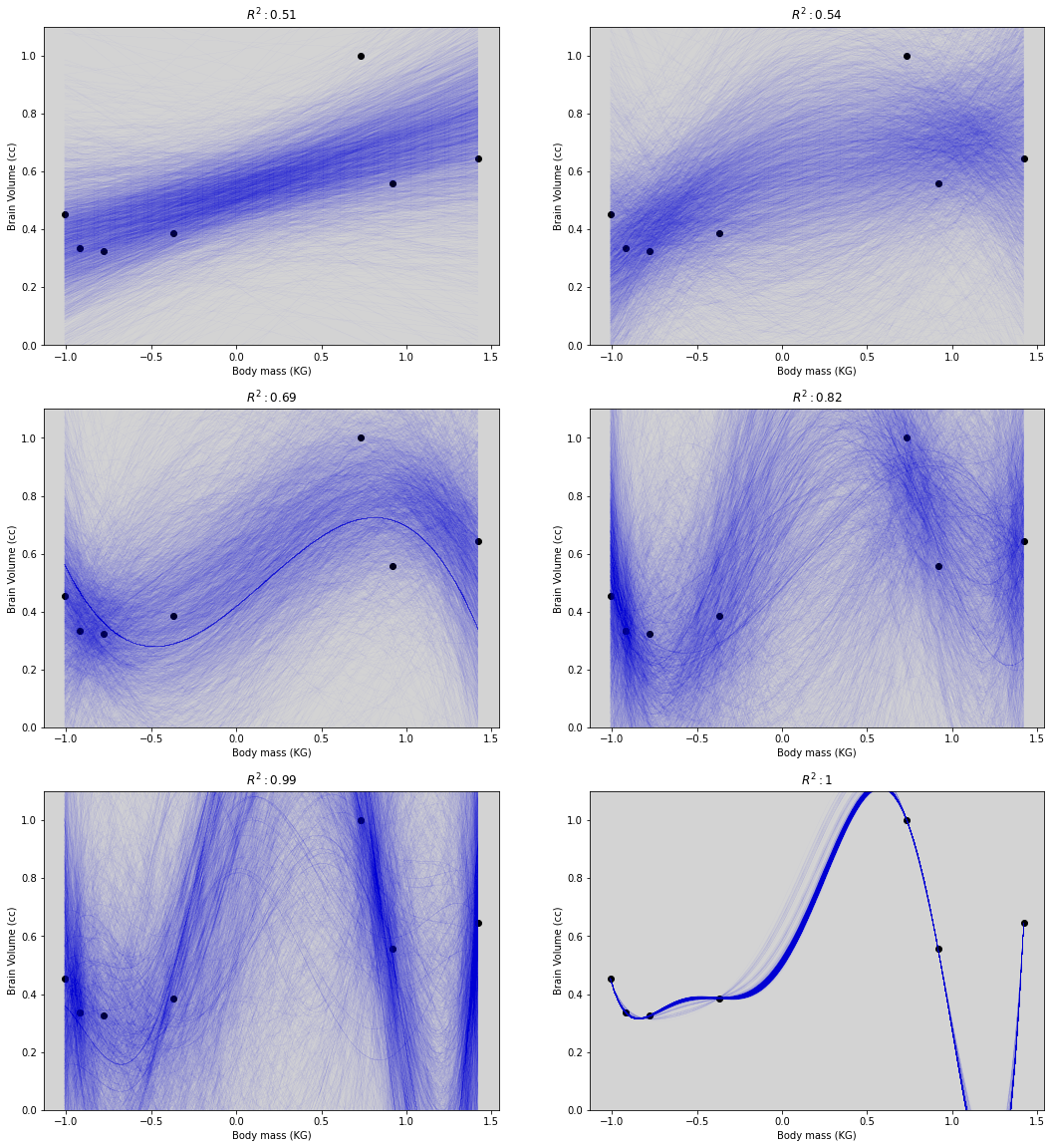
fig = plt.figure(figsize=(18, 20))
gs = GridSpec(nrows=3, ncols=2)
ax1 = fig.add_subplot(gs[0, 0])
ax1.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax1.plot(mass_seq, pp_1, c='blue', lw=0.01)
ax1.set_ylim(0, 1.1)
ax1.set_title('$R^2: 0.51$')
ax1.set_xlabel('Body mass (KG)')
ax1.set_ylabel('Brain Volume (cc)')
ax2 = fig.add_subplot(gs[0, 1])
ax2.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax2.plot(mass_seq, pp_2, c='blue', lw=0.01)
ax2.set_ylim(0, 1.1)
ax2.set_title('$R^2: 0.54$')
ax2.set_xlabel('Body mass (KG)')
ax2.set_ylabel('Brain Volume (cc)')
ax3 = fig.add_subplot(gs[1, 0])
ax3.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax3.plot(mass_seq, pp_3, c='blue', lw=0.01)
ax3.set_ylim(0, 1.1)
ax3.set_title('$R^2: 0.69$')
ax3.set_xlabel('Body mass (KG)')
ax3.set_ylabel('Brain Volume (cc)')
ax4 = fig.add_subplot(gs[1, 1])
ax4.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax4.plot(mass_seq, pp_4, c='blue', lw=0.01)
ax4.set_ylim(0, 1.1)
ax4.set_title('$R^2: 0.82$')
ax4.set_xlabel('Body mass (KG)')
ax4.set_ylabel('Brain Volume (cc)')
ax5 = fig.add_subplot(gs[2, 0])
ax5.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax5.plot(mass_seq, pp_5, c='blue', lw=0.01)
ax5.set_ylim(0, 1.1)
ax5.set_title('$R^2: 0.99$')
ax5.set_xlabel('Body mass (KG)')
ax5.set_ylabel('Brain Volume (cc)')
ax6 = fig.add_subplot(gs[2, 1])
ax6.scatter(d.mass_std.values, d.brain_std.values, c='black')
ax6.plot(mass_seq, pp_6, c='blue', lw=0.01)
ax6.set_ylim(0, 1.1)
ax6.set_title('$R^2: 1$')
ax6.set_xlabel('Body mass (KG)')
ax6.set_ylabel('Brain Volume (cc)')
plt.show()
R Code 7.11 - Pag 201¶
d.brain_std
0 0.324444
1 0.334815
2 0.453333
3 0.385926
4 0.557037
5 0.645185
6 1.000000
Name: brain_std, dtype: float64
Deletando a linha \(i=3\) do dataframe:
d_deleted_line_3 = d.brain_std.drop(3)
d_deleted_line_3
0 0.324444
1 0.334815
2 0.453333
4 0.557037
5 0.645185
6 1.000000
Name: brain_std, dtype: float64
R Code 7.12 - Pag 206¶
Entropia da informação: A incerteza contida na distribuição de probabilidade é a média do \(log\) da probabilidade do evento.
def H(p):
"""Information Entropy
H(p):= -sum(p_i * log(pi))
"""
if not np.sum(p) == 1:
print('ProbabilityError: This is not probability, its sum not equal to 1.')
else:
return - np.sum([p_i * np.log(p_i) if p_i > 0 else 0 for p_i in p])
p = (0.3, 0.7) # (p_rain, p_sum)
H(p)
0.6108643020548935
A incerteza do clima de Abu Dhabi é menor, pois é pouco provável que chova.
p_AbuDhabi = (0.01, 0.99) # (p_rain, p_sum)
H(p_AbuDhabi)
0.056001534354847345
p_3dim = (0.7, 0.15, 0.15) # (p_1, p_2, p_3)
H(p_3dim)
0.818808456222877
R Code 7.13 - pag 210¶
References:
Obs:
Here I learned to use the two blocks in stan: transformed parameters and generated quantities.
To calculate the Deviance it is necessary to make these changes.
That’s why I did all 6 estimates again with the calculations needed.
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta;
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta * body;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
beta ~ normal(0, 10);
sigma ~ lognormal(0, 1);
// Likelihood
brain ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] brain_hat;
for (i in 1:N){
log_lik[i] = normal_lpdf(brain[i] | mu[i], sigma);
brain_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_1 = stan.build(model, data=data)
samples_1 = posteriori_1.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[2];
real<lower=0> sigma;
}
transformed parameters{
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:2){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 10);
// Likelihood
brain ~ normal(mu, sigma);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_2 = stan.build(model, data=data)
samples_2 = posteriori_2.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[3];
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:3){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 10);
// Likelihood
brain ~ normal(mu, sigma);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_3 = stan.build(model, data=data)
samples_3 = posteriori_3.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[4];
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:4){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 10);
// Likelihood
brain ~ normal(mu, sigma);
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_4 = stan.build(model, data=data)
samples_4 = posteriori_4.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[5];
real<lower=0> sigma;
}
transformed parameters {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4 +
beta[5] * body^5;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:5){
beta[j] ~ normal(0, 10);
}
sigma ~ lognormal(0, 10);
// Likelihood
brain ~ normal(mu, sigma);
}
generated quantities {
vector[N] log_lik;
vector[N] brain_hat;
for (i in 1:N){
log_lik[i] = normal_lpdf(brain[i] | mu[i], sigma);
brain_hat[i] = normal_rng(mu[i], sigma);
}
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_5 = stan.build(model, data=data)
samples_5 = posteriori_5.sample(num_chains=4, num_samples=1000)
model = """
data {
int N;
vector[N] brain;
vector[N] body;
}
parameters {
real alpha;
real beta[6];
}
transformed parameters {
vector[N] mu;
mu = alpha + beta[1] * body +
beta[2] * body^2 +
beta[3] * body^3 +
beta[4] * body^4 +
beta[5] * body^5 +
beta[6] * body^6;
}
model {
// Prioris
alpha ~ normal(0.5, 1);
for (j in 1:6){
beta[j] ~ normal(0, 10);
}
// Likelihood
brain ~ normal(mu, 0.001);
}
generated quantities {
vector[N] log_ll_y;
for (i in 1:N){
log_ll_y[i] = normal_lpdf(brain[i] | mu, 0.001);
}
}
"""
data = {
'N': len(d.brain_std),
'brain': d.brain_std.values,
'body': d.mass_std.values,
}
posteriori_6 = stan.build(model, data=data)
samples_6 = posteriori_6.sample(num_chains=4, num_samples=1000)
def lppd(samples, outcome, std_residuals=True):
""" Calculate the LOG-POINTWISE-PREDICTIVE-DENSITY
samples : stan
Sampler results of fit posteriori.
Need 'mu' already computed.
outcome : list
List with outcomes the original data.
std_residuals : booleans
Compute lppd using std of the residuals
"""
mu = samples['mu']
N = len(outcome)
K = np.shape(mu)[1] # Qty samples (mu) sampled from posteriori
outcome = outcome.reshape(-1, 1)
if std_residuals:
sigma = (np.sum((mu - outcome)**2, 0) / N)**0.5
else:
sigma = samples['sigma'].flatten()
lppd = np.empty(N, dtype=float)
for i in range(N):
log_posteriori_predictive = stats.norm.logpdf(outcome[i], mu[i], sigma)
lppd[i] = np.log(np.sum(np.exp(log_posteriori_predictive))) - np.log(K)
return lppd
R Code 7.15¶
Like the book and the this one the values here have slight differeces.
Need review calculations - pag 211 - 2ed
lppd_lm = []
lppd_lm.append(np.sum(lppd(samples_1, d.brain_std.values, std_residuals=True)))
lppd_lm.append(np.sum(lppd(samples_2, d.brain_std.values, std_residuals=True)))
lppd_lm.append(np.sum(lppd(samples_3, d.brain_std.values, std_residuals=True)))
lppd_lm.append(np.sum(lppd(samples_4, d.brain_std.values, std_residuals=True)))
lppd_lm.append(np.sum(lppd(samples_5, d.brain_std.values, std_residuals=True)))
lppd_lm.append(np.sum(lppd(samples_6, d.brain_std.values, std_residuals=True)))
lppd_lm
[1.9566327201814353,
1.941588551424057,
2.14378226869432,
2.754228507821768,
5.480033073131333,
40.03033955975213]
import arviz as az
# ArviZ ships with style sheets!
# https://python.arviz.org/en/stable/examples/styles.html#example-styles
az.style.use("arviz-darkgrid")
data = az.from_pystan(
posterior=samples_5,
posterior_predictive="brain_hat",
observed_data=data,
log_likelihood={"brain": "log_lik"},
)
data
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, beta_dim_0: 5, mu_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 992 993 994 995 996 997 998 999 * beta_dim_0 (beta_dim_0) int64 0 1 2 3 4 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 Data variables: alpha (chain, draw) float64 0.7899 0.4723 0.1157 ... 0.7226 1.419 beta (chain, draw, beta_dim_0) float64 -1.853 -0.7106 ... 0.8986 sigma (chain, draw) float64 0.7009 0.3478 0.3989 ... 0.1128 0.1313 mu (chain, draw, mu_dim_0) float64 0.7289 0.2776 ... 0.533 1.005 Attributes: created_at: 2023-08-11T18:53:39.503098 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, brain_hat_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 * brain_hat_dim_0 (brain_hat_dim_0) int64 0 1 2 3 4 5 6 Data variables: brain_hat (chain, draw, brain_hat_dim_0) float64 -0.6716 ... 1.096 Attributes: created_at: 2023-08-11T18:53:39.659568 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, brain_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * brain_dim_0 (brain_dim_0) int64 0 1 2 3 4 5 6 Data variables: brain (chain, draw, brain_dim_0) float64 -0.7301 -0.5669 ... 1.111 Attributes: created_at: 2023-08-11T18:53:39.614761 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9942 0.9932 1.0 ... 0.7437 0.6009 step_size (chain, draw) float64 0.02283 0.02283 ... 0.01823 0.01823 tree_depth (chain, draw) int64 6 7 5 4 8 6 5 4 5 ... 5 6 7 7 5 7 8 7 7 n_steps (chain, draw) int64 63 159 31 31 255 ... 47 127 255 255 191 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 2.459 5.23 0.8842 ... -7.189 -5.73 lp (chain, draw) float64 -1.21 0.03107 2.038 ... 10.68 9.605 Attributes: created_at: 2023-08-11T18:53:39.561999 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0
data5 = az.from_pystan(
posterior=samples_5,
posterior_predictive="brain_hat",
observed_data=data,
log_likelihood={"brain": "log_lik"},
)
data5
-
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, beta_dim_0: 5, mu_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 992 993 994 995 996 997 998 999 * beta_dim_0 (beta_dim_0) int64 0 1 2 3 4 * mu_dim_0 (mu_dim_0) int64 0 1 2 3 4 5 6 Data variables: alpha (chain, draw) float64 0.7899 0.4723 0.1157 ... 0.7226 1.419 beta (chain, draw, beta_dim_0) float64 -1.853 -0.7106 ... 0.8986 sigma (chain, draw) float64 0.7009 0.3478 0.3989 ... 0.1128 0.1313 mu (chain, draw, mu_dim_0) float64 0.7289 0.2776 ... 0.533 1.005 Attributes: created_at: 2023-08-11T18:53:39.756266 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, brain_hat_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 * brain_hat_dim_0 (brain_hat_dim_0) int64 0 1 2 3 4 5 6 Data variables: brain_hat (chain, draw, brain_hat_dim_0) float64 -0.6716 ... 1.096 Attributes: created_at: 2023-08-11T18:53:39.902129 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000, brain_dim_0: 7) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * brain_dim_0 (brain_dim_0) int64 0 1 2 3 4 5 6 Data variables: brain (chain, draw, brain_dim_0) float64 -0.7301 -0.5669 ... 1.111 Attributes: created_at: 2023-08-11T18:53:39.855774 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 -
<xarray.Dataset> Dimensions: (chain: 4, draw: 1000) Coordinates: * chain (chain) int64 0 1 2 3 * draw (draw) int64 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 Data variables: acceptance_rate (chain, draw) float64 0.9942 0.9932 1.0 ... 0.7437 0.6009 step_size (chain, draw) float64 0.02283 0.02283 ... 0.01823 0.01823 tree_depth (chain, draw) int64 6 7 5 4 8 6 5 4 5 ... 5 6 7 7 5 7 8 7 7 n_steps (chain, draw) int64 63 159 31 31 255 ... 47 127 255 255 191 diverging (chain, draw) bool False False False ... False False False energy (chain, draw) float64 2.459 5.23 0.8842 ... -7.189 -5.73 lp (chain, draw) float64 -1.21 0.03107 2.038 ... 10.68 9.605 Attributes: created_at: 2023-08-11T18:53:39.807300 arviz_version: 0.15.1 inference_library: stan inference_library_version: 3.7.0 num_chains: 4 num_samples: 1000 num_thin: 1 num_warmup: 1000 save_warmup: 0
y_true = d.brain_std.values
y_pred = data5.posterior_predictive.stack(sample=("chain", "draw"))["brain_hat"].values.T
az.r2_score(y_true, y_pred)
r2 0.672509
r2_std 0.166309
dtype: float64
aa = az.loo(samples_5, pointwise=True)
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/stats.py:803: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.7 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
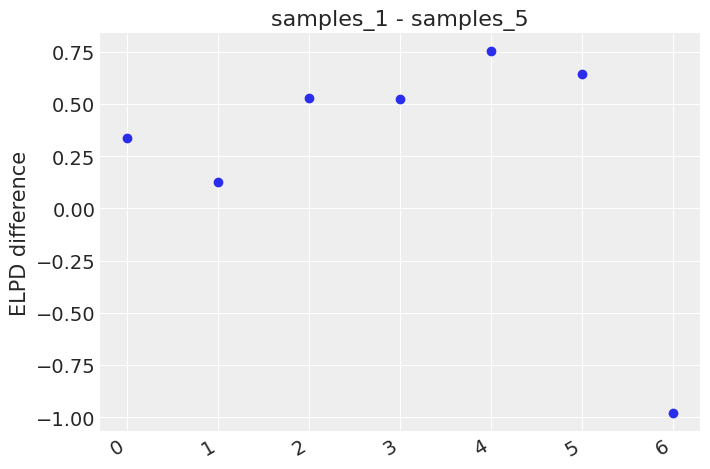
az.plot_elpd({'samples_1':samples_1, 'samples_5': samples_5}, xlabels=True)
plt.show()
/home/rodolpho/Projects/bayesian/BAYES/lib/python3.8/site-packages/arviz/stats/stats.py:803: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.7 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
aaa = data.posterior_predictive.brain_hat[0].values
np.log(np.sum(np.exp(aaa.T), 1)) - np.log(1000)
array([0.50019162, 0.61303674, 0.6133973 , 0.44845226, 0.79786415,
2.0166922 , 7.29542366])
R Code 7.16¶
Just using Rethinking Packages - pag 213
R Code 7.17¶
Just using Rethinking Packages - Pag 213